Geofold: topology-based protein unfolding pathways capture the effects of engineered disulfides on kinetic stability
- PMID: 22189917
- PMCID: PMC3277656
- DOI: 10.1002/prot.23249
Geofold: topology-based protein unfolding pathways capture the effects of engineered disulfides on kinetic stability
Abstract
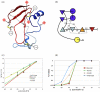
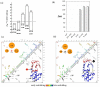
Protein unfolding is modeled as an ensemble of pathways, where each step in each pathway is the addition of one topologically possible conformational degree of freedom. Starting with a known protein structure, GeoFold hierarchically partitions (cuts) the native structure into substructures using revolute joints and translations. The energy of each cut and its activation barrier are calculated using buried solvent accessible surface area, side chain entropy, hydrogen bonding, buried cavities, and backbone degrees of freedom. A directed acyclic graph is constructed from the cuts, representing a network of simultaneous equilibria. Finite difference simulations on this graph simulate native unfolding pathways. Experimentally observed changes in the unfolding rates for disulfide mutants of barnase, T4 lysozyme, dihydrofolate reductase, and factor for inversion stimulation were qualitatively reproduced in these simulations. Detailed unfolding pathways for each case explain the effects of changes in the chain topology on the folding energy landscape. GeoFold is a useful tool for the inference of the effects of disulfide engineering on the energy landscape of protein unfolding.
Copyright © 2011 Wiley Periodicals, Inc.
Figures




Similar articles
-
Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities.Biochemistry. 1996 Apr 9;35(14):4298-305. doi: 10.1021/bi9524676. Biochemistry. 1996. PMID: 8605178
-
Engineered disulfide bonds as probes of the folding pathway of barnase: increasing the stability of proteins against the rate of denaturation.Biochemistry. 1993 Apr 27;32(16):4322-9. doi: 10.1021/bi00067a022. Biochemistry. 1993. PMID: 8476861
-
Stabilization of proteins by enhancement of inter-residue hydrophobic contacts: lessons of T4 lysozyme and barnase.J Biomol Struct Dyn. 2000 Dec;18(3):477-91. doi: 10.1080/07391102.2000.10506682. J Biomol Struct Dyn. 2000. PMID: 11149522
-
The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding.J Mol Biol. 1992 Apr 5;224(3):771-82. doi: 10.1016/0022-2836(92)90561-w. J Mol Biol. 1992. PMID: 1569556 Review.
-
The folding of an enzyme. III. Structure of the transition state for unfolding of barnase analysed by a protein engineering procedure.J Mol Biol. 1992 Apr 5;224(3):805-18. doi: 10.1016/0022-2836(92)90563-y. J Mol Biol. 1992. PMID: 1569558 Review.
Cited by
-
Anti-CTLA-4 nanobody as a promising approach in cancer immunotherapy.Cell Death Dis. 2024 Jan 8;15(1):17. doi: 10.1038/s41419-023-06391-x. Cell Death Dis. 2024. PMID: 38191571 Free PMC article. Review.
-
The structural basis of nanobody unfolding reversibility and thermoresistance.Sci Rep. 2018 May 21;8(1):7934. doi: 10.1038/s41598-018-26338-z. Sci Rep. 2018. PMID: 29784954 Free PMC article.
-
Mapping the Geometric Evolution of Protein Folding Motor.PLoS One. 2016 Oct 7;11(10):e0163993. doi: 10.1371/journal.pone.0163993. eCollection 2016. PLoS One. 2016. PMID: 27716851 Free PMC article.
-
Enhanced Thermostability of D-Psicose 3-Epimerase from Clostridium bolteae through Rational Design and Engineering of New Disulfide Bridges.Int J Mol Sci. 2021 Sep 16;22(18):10007. doi: 10.3390/ijms221810007. Int J Mol Sci. 2021. PMID: 34576170 Free PMC article.
-
Exploring the folding pathway of green fluorescent protein through disulfide engineering.Protein Sci. 2015 Mar;24(3):341-53. doi: 10.1002/pro.2621. Epub 2015 Jan 13. Protein Sci. 2015. PMID: 25516354 Free PMC article.
References
-
- Plaxco KWS, Baker KT, Contact Order D. Transition State Placement and the Refolding Rates of Single Domain Proteins. J Mol Biol. 1998;277(4):985–994. - PubMed
-
- Beck DA, Daggett V. Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods (San Diego, Calif. 2004;34(1):112–20. - PubMed
-
- Duan Y, Kollman PA. Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution. Science. 1998;282(5389):740–4. - PubMed
-
- Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4(1):10–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

