Hydroxyproline metabolism in mouse models of primary hyperoxaluria
- PMID: 22189945
- PMCID: PMC3311317
- DOI: 10.1152/ajprenal.00473.2011
Hydroxyproline metabolism in mouse models of primary hyperoxaluria
Abstract
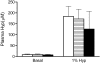
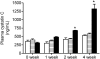
Primary hyperoxaluria type 1 (PH1) and type 2 (PH2) are rare genetic diseases that result from deficiencies in glyoxylate metabolism. The increased oxalate synthesis that occurs can lead to kidney stone formation, deposition of calcium oxalate in the kidney and other tissues, and renal failure. Hydroxyproline (Hyp) catabolism, which occurs mainly in the liver and kidney, is a prominent source of glyoxylate and could account for a significant portion of the oxalate produced in PH. To determine the sensitivity of mouse models of PH1 and PH2 to Hyp-derived oxalate, animals were fed diets containing 1% Hyp. Urinary excretions of glycolate and oxalate were used to monitor Hyp catabolism and the kidneys were examined to assess pathological changes. Both strains of knockout (KO) mice excreted more oxalate than wild-type (WT) animals with Hyp feeding. After 4 wk of Hyp feeding, all mice deficient in glyoxylate reductase/hydroxypyruvate reductase (GRHPR KO) developed severe nephrocalcinosis in contrast to animals deficient in alanine-glyoxylate aminotransferase (AGXT KO) where nephrocalcinosis was milder and with a lower frequency. Plasma cystatin C measurements over 4-wk Hyp feeding indicated no significant loss of renal function in WT and AGXT KO animals, and significant and severe loss of renal function in GRHPR KO animals after 2 and 4 wk, respectively. These data suggest that GRHPR activity may be vital in the kidney for limiting the conversion of Hyp-derived glyoxylate to oxalate. As Hyp catabolism may make a major contribution to the oxalate produced in PH patients, Hyp feeding in these mouse models should be useful in understanding the mechanisms associated with calcium oxalate deposition in the kidney.
Figures





Comment in
-
Re: hydroxyproline metabolism in mouse models of primary hyperoxaluria.J Urol. 2012 May;187(5):1928. doi: 10.1016/j.juro.2012.01.039. Epub 2012 Mar 21. J Urol. 2012. PMID: 22494766 No abstract available.
References
-
- Adams MR, Golden DL, Franke AA, Potter SM, Smith HS, Anthony MS. Dietary soy beta-conglycinin (7S globulin) inhibits atherosclerosis in mice. J Nutr 134: 511–516, 2004 - PubMed
-
- Cramer SD, Ferree PM, Lin K, Milliner DS, Holmes RP. The gene encoding hydroxypyruvate reductase (GRHPR) is mutated in patients with Primary Hyperoxaluria Type II. Hum Mol Genet 11: 2063–2069, 1999 - PubMed
-
- Danpure CJ. Primary hyperoxaluria. In: The Metabolic and Molecular Bases of Inherited Disease, edited by Scriver CR, Beaudet AL, Sly WS, Vallee D, Childs B, Kinzler KW, Vogelstein B. New York: McGraw-Hill, 2001, p. 3323–3367
-
- Frendeus KH, Wallin H, Janciauskiene S, Abrahamson M. Macrophage responses to interferon-gamma are dependent on cystatin C levels. Int J Biochem Cell Biol 41: 2262–2269, 2009 - PubMed
-
- Harris AH, Freel RW, Hatch M. Serum oxalate in human beings and rats as determined with the use of ion chromatography. J Lab Clin Med 144: 45–52, 2004 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

