Phylogeographical footprint of colonial history in the global dispersal of human immunodeficiency virus type 2 group A
- PMID: 22190015
- PMCID: PMC3542711
- DOI: 10.1099/vir.0.038638-0
Phylogeographical footprint of colonial history in the global dispersal of human immunodeficiency virus type 2 group A
Abstract
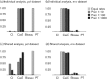
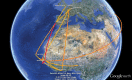
Human immunodeficiency virus type 2 (HIV-2) emerged in West Africa and has spread further to countries that share socio-historical ties with this region. However, viral origins and dispersal patterns at a global scale remain poorly understood. Here, we adopt a Bayesian phylogeographic approach to investigate the spatial dynamics of HIV-2 group A (HIV-2A) using a collection of 320 partial pol and 248 partial env sequences sampled throughout 19 countries worldwide. We extend phylogenetic diffusion models that simultaneously draw information from multiple loci to estimate location states throughout distinct phylogenies and explicitly attempt to incorporate human migratory fluxes. Our study highlights that Guinea-Bissau, together with Côte d'Ivoire and Senegal, have acted as the main viral sources in the early stages of the epidemic. We show that convenience sampling can obfuscate the estimation of the spatial root of HIV-2A. We explicitly attempt to circumvent this by incorporating rate priors that reflect the ratio of human flow from and to West Africa. We recover four main routes of HIV-2A dispersal that are laid out along colonial ties: Guinea-Bissau and Cape Verde to Portugal, Côte d'Ivoire and Senegal to France. Within Europe, we find strong support for epidemiological linkage from Portugal to Luxembourg and to the UK. We demonstrate that probabilistic models can uncover global patterns of HIV-2A dispersal providing sampling bias is taken into account and we provide a scenario for the international spread of this virus.
Figures


References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

