Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection
- PMID: 22190037
- PMCID: PMC3310910
- DOI: 10.1038/nature10693
Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection
Abstract
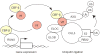
Restriction factors, such as the retroviral complementary DNA deaminase APOBEC3G, are cellular proteins that dominantly block virus replication. The AIDS virus, human immunodeficiency virus type 1 (HIV-1), produces the accessory factor Vif, which counteracts the host's antiviral defence by hijacking a ubiquitin ligase complex, containing CUL5, ELOC, ELOB and a RING-box protein, and targeting APOBEC3G for degradation. Here we reveal, using an affinity tag/purification mass spectrometry approach, that Vif additionally recruits the transcription cofactor CBF-β to this ubiquitin ligase complex. CBF-β, which normally functions in concert with RUNX DNA binding proteins, allows the reconstitution of a recombinant six-protein assembly that elicits specific polyubiquitination activity with APOBEC3G, but not the related deaminase APOBEC3A. Using RNA knockdown and genetic complementation studies, we also demonstrate that CBF-β is required for Vif-mediated degradation of APOBEC3G and therefore for preserving HIV-1 infectivity. Finally, simian immunodeficiency virus (SIV) Vif also binds to and requires CBF-β to degrade rhesus macaque APOBEC3G, indicating functional conservation. Methods of disrupting the CBF-β-Vif interaction might enable HIV-1 restriction and provide a supplement to current antiviral therapies that primarily target viral proteins.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


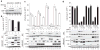
Comment in
-
HIV: Tagged for destruction.Nat Rev Microbiol. 2012 Jan 16;10(2):81. doi: 10.1038/nrmicro2739. Nat Rev Microbiol. 2012. PMID: 22245926 No abstract available.
References
-
- Goff SP. Retrovirus restriction factors. Mol Cell. 2004;16:849–859. - PubMed
-
- Malim MH, Emerman M. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. - PubMed
-
- Yu X, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI083196/AI/NIAID NIH HHS/United States
- P41 GM103481/GM/NIGMS NIH HHS/United States
- P41RR001614/RR/NCRR NIH HHS/United States
- P50GM081879/GM/NIGMS NIH HHS/United States
- P50 GM081879/GM/NIGMS NIH HHS/United States
- U54 RR022220/RR/NCRR NIH HHS/United States
- P50 GM082250/GM/NIGMS NIH HHS/United States
- P01 GM091743/GM/NIGMS NIH HHS/United States
- R01 AI064046/AI/NIAID NIH HHS/United States
- P41 RR001614/RR/NCRR NIH HHS/United States
- R01 GM078360/GM/NIGMS NIH HHS/United States
- P01 AI090935/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

