Seasonal abscisic acid signal and a basic leucine zipper transcription factor, DkbZIP5, regulate proanthocyanidin biosynthesis in persimmon fruit
- PMID: 22190340
- PMCID: PMC3271745
- DOI: 10.1104/pp.111.191205
Seasonal abscisic acid signal and a basic leucine zipper transcription factor, DkbZIP5, regulate proanthocyanidin biosynthesis in persimmon fruit
Abstract
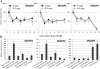
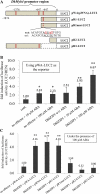
Proanthocyanidins (PAs) are secondary metabolites that contribute to plant protection and crop quality. Persimmon (Diospyros kaki) has a unique characteristic of accumulating large amounts of PAs, particularly in its fruit. Normal astringent-type and mutant nonastringent-type fruits show different PA accumulation patterns depending on the seasonal expression patterns of DkMyb4, which is a Myb transcription factor (TF) regulating many PA pathway genes in persimmon. In this study, attempts were made to identify the factors involved in DkMyb4 expression and the resultant PA accumulation in persimmon fruit. Treatment with abscisic acid (ABA) and an ABA biosynthesis inhibitor resulted in differential changes in the expression patterns of DkMyb4 and PA biosynthesis in astringent-type and nonastringent-type fruits depending on the development stage. To obtain an ABA-signaling TF, we isolated a full-length basic leucine zipper (bZIP) TF, DkbZIP5, which is highly expressed in persimmon fruit. We also showed that ectopic DkbZIP5 overexpression in persimmon calluses induced the up-regulation of DkMyb4 and the resultant PA biosynthesis. In addition, a detailed molecular characterization using the electrophoretic mobility shift assay and transient reporter assay indicated that DkbZIP5 recognized ABA-responsive elements in the promoter region of DkMyb4 and acted as a direct regulator of DkMyb4 in an ABA-dependent manner. These results suggest that ABA signals may be involved in PA biosynthesis in persimmon fruit via DkMyb4 activation by DkbZIP5.
Figures






References
-
- Akagi T, Ikegami A, Suzuki Y, Yoshida J, Yamada M, Sato A, Yonemori K. (2009a) Expression balances of structural genes in shikimate and flavonoid biosynthesis cause a difference in proanthocyanidin accumulation in persimmon (Diospyros kaki Thunb.) fruit. Planta 230: 899–915 - PubMed
-
- Akagi T, Ikegami A, Yonemori K. (2010a) DkMyb2 wound-induced transcription factor of persimmon (Diospyros kaki Thunb.), contributes to proanthocyanidin regulation. Planta 232: 1045–1059 - PubMed
-
- Akagi T, Kanzaki S, Gao M, Tao R, Parfitt DE, Yonemori K. (2009c) Quantitative real-time PCR to determine allele number for the astringency locus by analysis of a linked marker in Diospyros kaki Thunb. Tree Genet Genomes 5: 483–492
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

