Genetics of experimental allergic encephalomyelitis supports the role of T helper cells in multiple sclerosis pathogenesis
- PMID: 22190363
- PMCID: PMC4122509
- DOI: 10.1002/ana.22642
Genetics of experimental allergic encephalomyelitis supports the role of T helper cells in multiple sclerosis pathogenesis
Abstract
Objective: The major histocompatibility complex (MHC) is the primary genetic contributor to multiple sclerosis (MS) and experimental allergic encephalomyelitis (EAE), but multiple additional interacting loci are required for genetic susceptibility. The identity of most of these non-MHC genes is unknown. In this report, we identify genes within evolutionarily conserved genetic pathways leading to MS and EAE.
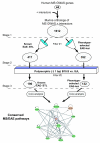
Methods: To identify non-MHC binary and quantitative trait loci (BTL/QTL) important in the pathogenesis of EAE, we generated phenotype-selected congenic mice using EAE-resistant B10.S and EAE-susceptible SJL mice. We hypothesized that genes linked to EAE BTL/QTL and MS-GWAS can be identified if they belong to common evolutionarily conserved pathways, which can be identified with a bioinformatic approach using Ingenuity software.
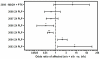
Results: Many known BTL/QTL were retained and linked to susceptibility during phenotype selection, the most significant being a region on chromosome 17 distal to H2 (Eae5). We show in pathway analysis that T helper (T(H))-cell differentiation genes are critical for both diseases. Bioinformatic analyses predicted that Eae5 is important in CD4 T-effector and/or Foxp3(+) T-regulatory cells (Tregs), and we found that B10.S-Eae5(SJL) congenic mice have significantly greater numbers of lymph node CD4 and Tregs than B10.S mice.
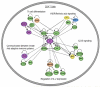
Interpretation: These results support the polygenic model of MS/EAE, whereby MHC and multiple minor loci are required for full susceptibility, and confirm a critical genetic dependence on CD4 T(H)-cell differentiation and function in the pathogenesis of both diseases.
Copyright © 2011 American Neurological Association.
Figures


Comment in
-
Decoding multiple sclerosis.Ann Neurol. 2011 Dec;70(6):A5-7. doi: 10.1002/ana.22680. Ann Neurol. 2011. PMID: 22190375 No abstract available.
References
-
- McFarland HF, Martin R. Multiple sclerosis: a complicated picture of autoimmunity. Nat.Immunol. 2007;8:913–919. - PubMed
-
- Schmidt H, Williamson D, Ashley-Koch A. HLA-DR15 haplotype and multiple sclerosis: a HuGE review. American journal of epidemiology. 2007;165:1097–1109. - PubMed
-
- Fazekas G, Tabira T. What transgenic and knockout mouse models teach us about experimental autoimmune encephalomyelitis. Rev.Immunogenet. 2000;2:115–132. - PubMed
-
- McDonald WI, Compston A, Edan G, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50:121–127. - PubMed
-
- Blankenhorn EP, Butterfield RJ, Rigby R, et al. Genetic analysis of the influence of pertussis toxin on experimental allergic encephalomyelitis susceptibility: an environmental agent can override genetic checkpoints. J Immunol. 2000;164:3420–3425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

