The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop
- PMID: 22190494
- PMCID: PMC3268300
- DOI: 10.1073/pnas.1105304109
The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop
Abstract
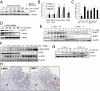
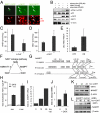
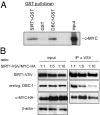
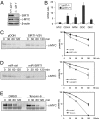
Silent information regulator 1 (SIRT1) represents an NAD(+)-dependent deacetylase that inhibits proapoptotic factors including p53. Here we determined whether SIRT1 is downstream of the prototypic c-MYC oncogene, which is activated in the majority of tumors. Elevated expression of c-MYC in human colorectal cancer correlated with increased SIRT1 protein levels. Activation of a conditional c-MYC allele induced increased levels of SIRT1 protein, NAD(+), and nicotinamide-phosphoribosyltransferase (NAMPT) mRNA in several cell types. This increase in SIRT1 required the induction of the NAMPT gene by c-MYC. NAMPT is the rate-limiting enzyme of the NAD(+) salvage pathway and enhances SIRT1 activity by increasing the amount of NAD(+). c-MYC also contributed to SIRT1 activation by sequestering the SIRT1 inhibitor deleted in breast cancer 1 (DBC1) from the SIRT1 protein. In primary human fibroblasts previously immortalized by introduction of c-MYC, down-regulation of SIRT1 induced senescence and apoptosis. In various cell lines inactivation of SIRT1 by RNA interference, chemical inhibitors, or ectopic DBC1 enhanced c-MYC-induced apoptosis. Furthermore, SIRT1 directly bound to and deacetylated c-MYC. Enforced SIRT1 expression increased and depletion/inhibition of SIRT1 reduced c-MYC stability. Depletion/inhibition of SIRT1 correlated with reduced lysine 63-linked polyubiquitination of c-Myc, which presumably destabilizes c-MYC by supporting degradative lysine 48-linked polyubiquitination. Moreover, SIRT1 enhanced the transcriptional activity of c-MYC. Taken together, these results show that c-MYC activates SIRT1, which in turn promotes c-MYC function. Furthermore, SIRT1 suppressed cellular senescence in cells with deregulated c-MYC expression and also inhibited c-MYC-induced apoptosis. Constitutive activation of this positive feedback loop may contribute to the development and maintenance of tumors in the context of deregulated c-MYC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



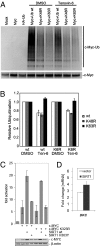
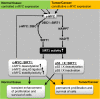
References
-
- Secombe J, Pierce SB, Eisenman RN. Myc: A weapon of mass destruction. Cell. 2004;117:153–156. - PubMed
-
- Dang CV, et al. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. - PubMed
-
- Hermeking H. The MYC oncogene as a cancer drug target. Curr Cancer Drug Targets. 2003;3:163–175. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

