Genome-scale phylogeny of the alphavirus genus suggests a marine origin
- PMID: 22190718
- PMCID: PMC3302268
- DOI: 10.1128/JVI.05591-11
Genome-scale phylogeny of the alphavirus genus suggests a marine origin
Abstract
The genus Alphavirus comprises a diverse group of viruses, including some that cause severe disease. Using full-length sequences of all known alphaviruses, we produced a robust and comprehensive phylogeny of the Alphavirus genus, presenting a more complete evolutionary history of these viruses compared to previous studies based on partial sequences. Our phylogeny suggests the origin of the alphaviruses occurred in the southern oceans and spread equally through the Old and New World. Since lice appear to be involved in aquatic alphavirus transmission, it is possible that we are missing a louse-borne branch of the alphaviruses. Complete genome sequencing of all members of the genus also revealed conserved residues forming the structural basis of the E1 and E2 protein dimers.
Figures



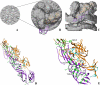
References
-
- Anderson CR, Downs WG, Wattley GH, Ahin NW, Reese AA. 1957. Mayaro virus: a new human disease agent. II. Isolation from blood of patients in Trinidad, B. W. I. Am. J. Trop. Med. Hyg. 6:1012–1016 - PubMed
-
- Bigler WJ, Ventura AK, Lewis AL, Wellings FM, Ehrenkanz NJ. 1974. Venezuelan equine encephalomyelitis in Florida: endemic virus circulation in native rodent populations of Everglades hammocks. Am. J. Trop. Med. Hyg. 23:513–521 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

