Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages
- PMID: 22190722
- PMCID: PMC3302286
- DOI: 10.1128/JVI.06157-11
Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages
Abstract
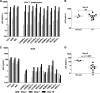
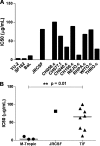
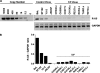
Genome sequences of transmitted/founder (T/F) HIV-1 have been inferred by analyzing single genome amplicons of acute infection plasma viral RNA in the context of a mathematical model of random virus evolution; however, few of these T/F sequences have been molecularly cloned and biologically characterized. Here, we describe the derivation and biological analysis of ten infectious molecular clones, each representing a T/F genome responsible for productive HIV-1 clade B clinical infection. Each of the T/F viruses primarily utilized the CCR5 coreceptor for entry and replicated efficiently in primary human CD4(+) T lymphocytes. This result supports the conclusion that single genome amplification-derived sequences from acute infection allow for the inference of T/F viral genomes that are consistently replication competent. Studies with monocyte-derived macrophages (MDM) demonstrated various levels of replication among the T/F viruses. Although all T/F viruses replicated in MDM, the overall replication efficiency was significantly lower compared to prototypic "highly macrophage-tropic" virus strains. This phenotype was transferable by expressing the env genes in an isogenic proviral DNA backbone, indicating that T/F virus macrophage tropism mapped to Env. Furthermore, significantly higher concentrations of soluble CD4 were required to inhibit T/F virus infection compared to prototypic macrophage-tropic virus strains. Our findings suggest that the acquisition of clinical HIV-1 subtype B infection occurs by mucosal exposure to virus that is not highly macrophage tropic and that the generation and initial biological characterization of 10 clade B T/F infectious molecular clones provides new opportunities to probe virus-host interactions involved in HIV-1 transmission.
Figures

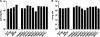




References
-
- Alkhatib G, et al. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958 - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

