Analysis of yeast endocytic site formation and maturation through a regulatory transition point
- PMID: 22190733
- PMCID: PMC3279393
- DOI: 10.1091/mbc.E11-02-0108
Analysis of yeast endocytic site formation and maturation through a regulatory transition point
Erratum in
- Mol Biol Cell. 2012 Aug;23(16):3275
Abstract
The earliest stages of endocytic site formation and the regulation of endocytic site maturation are not well understood. Here we analyzed the order in which the earliest proteins are detectable at endocytic sites in budding yeast and found that an uncharacterized protein, Pal1p/Ydr348cp, is also present at the initial stages of endocytosis. Because Ede1p (homologue of Eps15) and clathrin are the early-arriving proteins most important for cargo uptake, their roles during the early stages of endocytosis were examined more comprehensively. Ede1p is necessary for efficient recruitment of most early-arriving proteins, but not for the recruitment of the adaptor protein Yap1802p, to endocytic sites. The early-arriving proteins, as well as the later-arriving proteins Sla2p and Ent1/2p (homologues of Hip1R and epsins), were found to have longer lifetimes in CLC1-knockout yeast, which indicates that clathrin light chain facilitates the transition from the intermediate to late coat stages. Cargo also arrives during the early stages of endocytosis, and therefore its effect on endocytic machinery dynamics was investigated. Our results are consistent with a role for cargo in regulating the transition of endocytic sites from the early stages of formation to the late stages during which vesicle formation occurs.
Figures


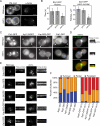
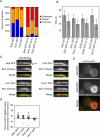

References
-
- Baggett JJ, Shaw JD, Sciambi CJ, Watson HA, Wendland B. Fluorescent labeling of yeast. Curr Protoc Cell Biol. 2003;Chapter 14 Unit 4.13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

