LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis
- PMID: 22190735
- PMCID: PMC3279398
- DOI: 10.1091/mbc.E11-06-0530
LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis
Abstract
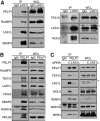
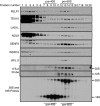
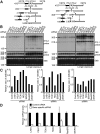
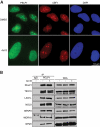
The coordination of RNA polymerase I transcription with pre-rRNA processing, preribosomal particle assembly, and nuclear export is a finely tuned process requiring the concerted actions of a number of accessory factors. However, the exact functions of some of these proteins and how they assemble in subcomplexes remain poorly defined. LAS1L was first described as a nucleolar protein required for maturation of the 60S preribosomal subunit. In this paper, we demonstrate that LAS1L interacts with PELP1, TEX10, and WDR18, the mammalian homologues of the budding yeast Rix1 complex, along with NOL9 and SENP3, to form a novel nucleolar complex that cofractionates with the 60S preribosomal subunit. Depletion of LAS1L-associated proteins results in a p53-dependent G1 arrest and leads to defects in processing of the pre-rRNA internal transcribed spacer 2 region. We further show that the nucleolar localization of this complex requires active RNA polymerase I transcription and the small ubiquitin-like modifier-specific protease SENP3. Taken together, our data identify a novel mammalian complex required for 60S ribosomal subunit synthesis, providing further insight into the intricate, yet poorly described, process of ribosome biogenesis in higher eukaryotes.
Figures






References
-
- Arabi A, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. - PubMed
-
- Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell. 2001;8:517–529. - PubMed
-
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. - PubMed
-
- Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–1124. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

