Characterization of the stop codon readthrough signal of Colorado tick fever virus segment 9 RNA
- PMID: 22190746
- PMCID: PMC3264911
- DOI: 10.1261/rna.030338.111
Characterization of the stop codon readthrough signal of Colorado tick fever virus segment 9 RNA
Abstract
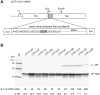
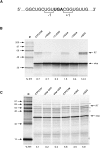
Termination codon readthrough is utilized as a mechanism of expression of a growing number of viral and cellular proteins, but in many cases the mRNA signals that promote readthrough are poorly characterized. Here, we investigated the readthrough signal of Colorado tick fever virus (CTFV) segment 9 RNA (Seg-9). CTFV is the type-species of the genus Coltivirus within the family Reoviridae and is a tick-borne, double-stranded, segmented RNA virus. Seg-9 encodes a 36-kDa protein VP9, and by readthrough of a UGA stop codon, a 65-kDa product, VP9'. Using a reporter system, we defined the minimal sequence requirements for readthrough and confirmed activity in both mammalian and insect cell-free translation systems, and in transfected mammalian cells. Mutational analysis revealed that readthrough was UGA specific, and that the local sequence context around the UGA influenced readthrough efficiency. Readthrough was also dependent upon a stable RNA stem-loop structure beginning eight bases downstream from the UGA codon. Mutational analysis of this stem-loop revealed a requirement for the stem region but not for substructures identified within the loop. Unexpectedly, we were unable to detect a ribosomal pause during translation of the CTFV signal, suggesting that the mechanism of readthrough, at least at this site, is unlikely to be dependent upon RNA secondary-structure induced ribosomal pausing at the recoded stop codon.
Figures




References
-
- Alam SL, Wills NM, Ingram JA, Atkins JF, Gesteland RF 1999. Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus. J Mol Biol 288: 837–852 - PubMed
-
- Arkov AL, Murgola EJ 1999. Ribosomal RNAs in translation termination: facts and hypotheses. Biochemistry (Mosc) 64: 1354–1359 - PubMed
-
- Bertram G, Innes S, Minella O, Richardson JP, Stansfield I 2001. Endless possibilities: translation termination and stop codon recognition. Microbiology 147: 255–269 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
