An integrated framework to model cellular phenotype as a component of biochemical networks
- PMID: 22190923
- PMCID: PMC3235418
- DOI: 10.1155/2011/608295
An integrated framework to model cellular phenotype as a component of biochemical networks
Abstract
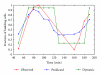
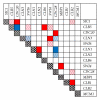
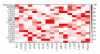
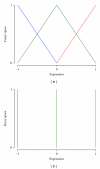
Identification of regulatory molecules in signaling pathways is critical for understanding cellular behavior. Given the complexity of the transcriptional gene network, the relationship between molecular expression and phenotype is difficult to determine using reductionist experimental methods. Computational models provide the means to characterize regulatory mechanisms and predict phenotype in the context of gene networks. Integrating gene expression data with phenotypic data in transcriptional network models enables systematic identification of critical molecules in a biological network. We developed an approach based on fuzzy logic to model cell budding in Saccharomyces cerevisiae using time series expression microarray data of the cell cycle. Cell budding is a phenotype of viable cells undergoing division. Predicted interactions between gene expression and phenotype reflected known biological relationships. Dynamic simulation analysis reproduced the behavior of the yeast cell cycle and accurately identified genes and interactions which are essential for cell viability.
Figures

References
-
- Hautaniemi S, Kharait S, Iwabu A, Wells A, Lauffenburger DA. Modeling of signal-response cascades using decision tree analysis. Bioinformatics. 2005;21(9):2027–2035. - PubMed
-
- Asthagiri AR, Lauffenburger DA. Bioengineering models of cell signaling. Annual Review of Biomedical Engineering. 2000;2(2000):31–53. - PubMed
-
- Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004;429(6987):92–96. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

