Evaluation of front-end higher energy collision-induced dissociation on a benchtop dual-pressure linear ion trap mass spectrometer for shotgun proteomics
- PMID: 22192247
- PMCID: PMC3277647
- DOI: 10.1021/ac203210a
Evaluation of front-end higher energy collision-induced dissociation on a benchtop dual-pressure linear ion trap mass spectrometer for shotgun proteomics
Abstract
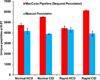
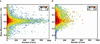
We report the implementation of front-end higher energy collision-induced dissociation (fHCD) on a benchtop dual-pressure linear ion trap. Software and hardware modifications were employed, described in detail vide-infra, to allow isolated ions to undergo collisions with ambient gas molecules in an intermediate multipole (q00) of the instrument. Results comparing the performance of fHCD and resonance excitation collision-induced dissociation (RE-CID) in terms of injection time, total number of scans, efficiency, mass measurement accuracy (MMA), unique peptide identifications, and spectral quality of labile modified peptides are presented. fHCD is approximately 23% as efficient as RE-CID, and depending on the search algorithm, it identifies 6.6% more or 15% less peptides (q < 0.01) from a soluble whole-cell lysate ( Caenorhabditis elegans ) than RE-CID using Mascot or Sequest search algorithms, respectively. fHCD offers a clear advantage for the analysis of phosphorylated and glycosylated (O-GlcNAc) peptides as the average cross-correlation score (XCorr) for spectra using fHCD was statistically greater (p < 0.05) than for spectra collected using RE-CID.
© 2011 American Chemical Society
Figures




Similar articles
-
Identification of proteins and phosphoproteins using pulsed Q collision induced dissociation (PQD).J Am Soc Mass Spectrom. 2011 Oct;22(10):1753-62. doi: 10.1007/s13361-011-0197-6. Epub 2011 Jul 15. J Am Soc Mass Spectrom. 2011. PMID: 21952889 Free PMC article.
-
A new ion mobility-linear ion trap instrument for complex mixture analysis.Anal Chem. 2014 Aug 19;86(16):8121-8. doi: 10.1021/ac501527y. Epub 2014 Aug 6. Anal Chem. 2014. PMID: 25068446 Free PMC article.
-
A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed.Mol Cell Proteomics. 2009 Dec;8(12):2759-69. doi: 10.1074/mcp.M900375-MCP200. Epub 2009 Oct 14. Mol Cell Proteomics. 2009. PMID: 19828875 Free PMC article.
-
Collision-induced dissociation (CID) of peptides and proteins.Methods Enzymol. 2005;402:148-85. doi: 10.1016/S0076-6879(05)02005-7. Methods Enzymol. 2005. PMID: 16401509 Review.
-
[Recent advances in glycopeptide enrichment and mass spectrometry data interpretation approaches for glycoproteomics analyses].Se Pu. 2021 Oct;39(10):1045-1054. doi: 10.3724/SP.J.1123.2021.06011. Se Pu. 2021. PMID: 34505426 Free PMC article. Review. Chinese.
Cited by
-
Glycoprotein Enrichment Analytical Techniques: Advantages and Disadvantages.Methods Enzymol. 2017;585:397-429. doi: 10.1016/bs.mie.2016.11.009. Epub 2017 Jan 16. Methods Enzymol. 2017. PMID: 28109440 Free PMC article.
-
Neutron encoded labeling for peptide identification.Anal Chem. 2013 May 21;85(10):5129-37. doi: 10.1021/ac400476w. Epub 2013 May 2. Anal Chem. 2013. PMID: 23638792 Free PMC article.
-
Toxicoproteomic analysis of pulmonary carbon nanotube exposure using LC-MS/MS.Toxicology. 2015 Mar 2;329:80-7. doi: 10.1016/j.tox.2015.01.011. Epub 2015 Jan 15. Toxicology. 2015. PMID: 25598225 Free PMC article.
-
Discrimination of Isomers of Released N- and O-Glycans Using Diagnostic Product Ions in Negative Ion PGC-LC-ESI-MS/MS.J Am Soc Mass Spectrom. 2018 Jun;29(6):1194-1209. doi: 10.1007/s13361-018-1932-z. Epub 2018 Mar 30. J Am Soc Mass Spectrom. 2018. PMID: 29603058
-
Learning score function parameters for improved spectrum identification in tandem mass spectrometry experiments.J Proteome Res. 2012 Sep 7;11(9):4499-508. doi: 10.1021/pr300234m. Epub 2012 Aug 15. J Proteome Res. 2012. PMID: 22866926 Free PMC article.
References
-
- Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Nat Methods. 2007;4:923–925. - PubMed
-
- Syka JEP, Marto JA, Bai DL, Horning S, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, Shabanowitz J, Hunt DF. J Proteome Res. 2004;3:621–626. - PubMed
-
- Hu QZ, Noll RJ, Li HY, Makarov A, Hardman M, Cooks RG. J Mass Spectrom. 2005;40:430–443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

