Bioinformatic characterization of the 4-Toluene Sulfonate Uptake Permease (TSUP) family of transmembrane proteins
- PMID: 22192777
- PMCID: PMC3917603
- DOI: 10.1016/j.bbamem.2011.12.005
Bioinformatic characterization of the 4-Toluene Sulfonate Uptake Permease (TSUP) family of transmembrane proteins
Abstract
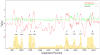
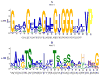
The ubiquitous sequence diverse 4-Toluene Sulfonate Uptake Permease (TSUP) family contains few characterized members and is believed to catalyze the transport of several sulfur-based compounds. Prokaryotic members of the TSUP family outnumber the eukaryotic members substantially, and in prokaryotes, but not eukaryotes, extensive lateral gene transfer occurred during family evolution. Despite unequal representation, homologues from the three taxonomic domains of life share well-conserved motifs. We show that the prototypical eight TMS topology arose from an intragenic duplication of a four transmembrane segment (TMS) unit. Possibly, a two TMS α-helical hairpin structure was the precursor of the 4 TMS repeat unit. Genome context analyses confirmed the proposal of a sulfur-based compound transport role for many TSUP homologues, but functional outliers appear to be prevalent as well. Preliminary results suggest that the TSUP family is a member of a large novel superfamily that includes rhodopsins, integral membrane chaperone proteins, transmembrane electron flow carriers and several transporter families. All of these proteins probably arose via the same pathway: 2→4→8 TMSs followed by loss of a TMS either at the N- or C-terminus, depending on the family, to give the more frequent 7 TMS topology.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures






References
-
- Busch W, Saier MH., Jr The transporter classification (TC) system, 2002. Crit Rev Biochem Mol Biol. 2002;37:287–337. - PubMed
-
- Gatti L, Cossa G, Beretta GL, Zaffaroni N, Perego P. Novel Insights into Targeting ATP-Binding Cassette Transporters for Antitumor Therapy. Current Medicinal Chemistry. 2011 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

