Use of parallel validation high-throughput screens to reduce false positives and identify novel dengue NS2B-NS3 protease inhibitors
- PMID: 22193283
- PMCID: PMC3266433
- DOI: 10.1016/j.antiviral.2011.12.003
Use of parallel validation high-throughput screens to reduce false positives and identify novel dengue NS2B-NS3 protease inhibitors
Abstract
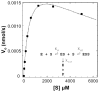
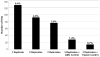
Dengue virus (DENV), a mosquito-borne member of the family Flaviviridae, is a significant global pathogen affecting primarily tropical and subtropical regions of the world and placing tremendous burden on the limited medical infrastructure that exists in many of the developing countries located within these regions. Recent outbreaks in developed countries, including Australia (Hanna et al., 2009), France (La Ruche et al., 2010), Taiwan (Kuan et al., 2010), and the USA (CDC, 2010), lead many researchers to believe that continued emergence into more temperate latitudes is likely. A primary concern is that there are no approved vaccines or antiviral therapies to treat DENV infections. Since the viral NS2B-NS3 protease (DENV NS2B-NS3pro) is required for virus replication, it provides a strategic target for the development of antiviral drugs. In this study, proof-of-concept high-throughput screenings (HTSs) were performed to unambiguously identify dengue 2 virus (DEN2V) NS2B-NS3pro inhibitors from a library of 2000 compounds. Validation screens were performed in parallel to concurrently eliminate insoluble, auto-fluorescing, and/or nonspecific inhibitors. Kinetic analyses of the hits revealed that parallel substrate fluorophore (AMC) interference controls and trypsin inhibition controls were able to reduce false positive rates due to solubility and fluorophore interference while the trypsin inhibition control additionally eliminated non-specific inhibitors. We identified five DEN2V NS2B-NS3pro inhibitors that also inhibited the related West Nile virus (WNV) protease (NS2B-NS3pro), but did not inhibit the trypsin protease. Biochemical analyses revealed various mechanisms of inhibition including competitive and mixed noncompetitive inhibition, with the lowest K(i) values being 12±1.5 μM for DEN2V NS2B-NS3pro and 2±0.2 μM for WNV NS2B-NS3pro.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures
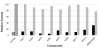



References
-
- Alvarez M, Rodriguez-Roche R, Bernardo L, Vazquez S, Morier L, Gonzalez D, Castro O, Kouri G, Halstead SB, Guzman MG. Dengue hemorrhagic fever caused by sequential dengue 1–3 virus infections over a long time interval: Havana epidemic, 2001–2002. Am J Trop Med Hyg. 2006;75:1113–1117. - PubMed
-
- CDC . Locally acquired Dengue--Key West, Florida, 2009–2010. MMWR Morb Mortal Wkly Rep. 2010;59:577–581. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

