Comprehensive qPCR profiling of gene expression in single neuronal cells
- PMID: 22193304
- PMCID: PMC4377330
- DOI: 10.1038/nprot.2011.430
Comprehensive qPCR profiling of gene expression in single neuronal cells
Abstract
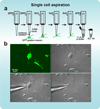
A major challenge in neuronal stem cell biology lies in characterization of lineage-specific reprogrammed human neuronal cells, a process that necessitates the use of an assay sensitive to the single-cell level. Single-cell gene profiling can provide definitive evidence regarding the conversion of one cell type into another at a high level of resolution. The protocol we describe uses Fluidigm Biomark dynamic arrays for high-throughput expression profiling from single neuronal cells, assaying up to 96 independent samples with up to 96 quantitative PCR (qPCR) probes (equivalent to 9,216 reactions) in a single experiment, which can be completed within 2-3 d. The protocol enables simple and cost-effective profiling of several hundred transcripts from a single cell, and it could have numerous utilities.
Figures


References
-
- Li HH, et al. Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature. 1988;335(6189):414. - PubMed
-
- Cauli Bruno, Lambolez Bertrand. Unravelling Single Cell Genomics. The Royal Society of Chemistry; 2010. p. 81.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

