Accurate, rapid taxonomic classification of fungal large-subunit rRNA genes
- PMID: 22194300
- PMCID: PMC3294464
- DOI: 10.1128/AEM.06826-11
Accurate, rapid taxonomic classification of fungal large-subunit rRNA genes
Abstract
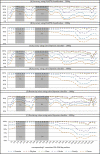
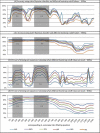
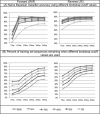
Taxonomic and phylogenetic fingerprinting based on sequence analysis of gene fragments from the large-subunit rRNA (LSU) gene or the internal transcribed spacer (ITS) region is becoming an integral part of fungal classification. The lack of an accurate and robust classification tool trained by a validated sequence database for taxonomic placement of fungal LSU genes is a severe limitation in taxonomic analysis of fungal isolates or large data sets obtained from environmental surveys. Using a hand-curated set of 8,506 fungal LSU gene fragments, we determined the performance characteristics of a naïve Bayesian classifier across multiple taxonomic levels and compared the classifier performance to that of a sequence similarity-based (BLASTN) approach. The naïve Bayesian classifier was computationally more rapid (>460-fold with our system) than the BLASTN approach, and it provided equal or superior classification accuracy. Classifier accuracies were compared using sequence fragments of 100 bp and 400 bp and two different PCR primer anchor points to mimic sequence read lengths commonly obtained using current high-throughput sequencing technologies. Accuracy was higher with 400-bp sequence reads than with 100-bp reads. It was also significantly affected by sequence location across the 1,400-bp test region. The highest accuracy was obtained across either the D1 or D2 variable region. The naïve Bayesian classifier provides an effective and rapid means to classify fungal LSU sequences from large environmental surveys. The training set and tool are publicly available through the Ribosomal Database Project.
Figures



References
-
- Arnold AE, et al. 2009. A phylogenetic estimation of trophic transition networks for ascomycetous fungi: are lichens cradles of symbiotrophic fungal diversification? Syst. Biol. 58:283–297 - PubMed
-
- Blackwell M. 2011. The fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 98:426–438 - PubMed
-
- Blackwell M, Hibbett DS, Taylor JW, Spatafora JW. 2006. Research coordination networks: a phylogeny for kingdom Fungi (deep hypha). Mycologia 98:829–837 - PubMed
-
- Buee M, et al. 2009. 454 pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol. 184:449–456 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

