Widespread genetic switches and toxicity resistance proteins for fluoride
- PMID: 22194412
- PMCID: PMC4140402
- DOI: 10.1126/science.1215063
Widespread genetic switches and toxicity resistance proteins for fluoride
Abstract
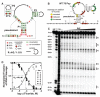
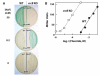
Most riboswitches are metabolite-binding RNA structures located in bacterial messenger RNAs where they control gene expression. We have discovered a riboswitch class in many bacterial and archaeal species whose members are selectively triggered by fluoride but reject other small anions, including chloride. These fluoride riboswitches activate expression of genes that encode putative fluoride transporters, enzymes that are known to be inhibited by fluoride, and additional proteins of unknown function. Our findings indicate that most organisms are naturally exposed to toxic levels of fluoride and that many species use fluoride-sensing RNAs to control the expression of proteins that alleviate the deleterious effects of this anion.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

