A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing
- PMID: 22194472
- PMCID: PMC3290779
- DOI: 10.1101/gr.133645.111
A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing
Abstract
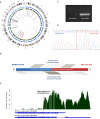
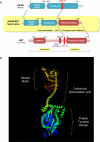
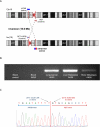
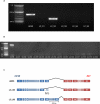
The identification of the molecular events that drive cancer transformation is essential to the development of targeted agents that improve the clinical outcome of lung cancer. Many studies have reported genomic driver mutations in non-small-cell lung cancers (NSCLCs) over the past decade; however, the molecular pathogenesis of >40% of NSCLCs is still unknown. To identify new molecular targets in NSCLCs, we performed the combined analysis of massively parallel whole-genome and transcriptome sequencing for cancer and paired normal tissue of a 33-yr-old lung adenocarcinoma patient, who is a never-smoker and has no familial cancer history. The cancer showed no known driver mutation in EGFR or KRAS and no EML4-ALK fusion. Here we report a novel fusion gene between KIF5B and the RET proto-oncogene caused by a pericentric inversion of 10p11.22-q11.21. This fusion gene overexpresses chimeric RET receptor tyrosine kinase, which could spontaneously induce cellular transformation. We identified the KIF5B-RET fusion in two more cases out of 20 primary lung adenocarcinomas in the replication study. Our data demonstrate that a subset of NSCLCs could be caused by a fusion of KIF5B and RET, and suggest the chimeric oncogene as a promising molecular target for the personalized diagnosis and treatment of lung cancer.
Figures

References
-
- Alberti L, Carniti C, Miranda C, Roccato E, Pierotti MA 2003. RET and NTRK1 proto-oncogenes in human diseases. J Cell Physiol 195: 168–186 - PubMed
-
- Daire V, Pous C 2011. Kinesins and protein kinases: key players in the regulation of microtubule dynamics and organization. Arch Biochem Biophys 510: 83–92 - PubMed
-
- Dawson DM, Lawrence EG, MacLennan GT, Amini SB, Kung HJ, Robinson D, Resnick MI, Kursh ED, Pretlow TP, Pretlow TG 1998. Altered expression of RET proto-oncogene product in prostatic intraepithelial neoplasia and prostate cancer. J Natl Cancer Inst 90: 519–523 - PubMed
-
- Durbec P, Marcos-Gutierrez CV, Kilkenny C, Grigoriou M, Wartiowaara K, Suvanto P, Smith D, Ponder B, Costantini F, Saarma M, et al. 1996. GDNF signalling through the Ret receptor tyrosine kinase. Nature 381: 789–793 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
