The ribosome modulates nascent protein folding
- PMID: 22194581
- PMCID: PMC4172366
- DOI: 10.1126/science.1209740
The ribosome modulates nascent protein folding
Abstract
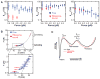
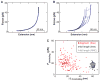
Proteins are synthesized by the ribosome and generally must fold to become functionally active. Although it is commonly assumed that the ribosome affects the folding process, this idea has been extremely difficult to demonstrate. We have developed an experimental system to investigate the folding of single ribosome-bound stalled nascent polypeptides with optical tweezers. In T4 lysozyme, synthesized in a reconstituted in vitro translation system, the ribosome slows the formation of stable tertiary interactions and the attainment of the native state relative to the free protein. Incomplete T4 lysozyme polypeptides misfold and aggregate when free in solution, but they remain folding-competent near the ribosomal surface. Altogether, our results suggest that the ribosome not only decodes the genetic information and synthesizes polypeptides, but also promotes efficient de novo attainment of the native state.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

