Evolution of microRNA genes in Oryza sativa and Arabidopsis thaliana: an update of the inverted duplication model
- PMID: 22194805
- PMCID: PMC3237417
- DOI: 10.1371/journal.pone.0028073
Evolution of microRNA genes in Oryza sativa and Arabidopsis thaliana: an update of the inverted duplication model
Abstract
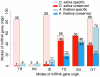
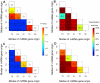
The origin and evolution of microRNA (miRNA) genes, which are of significance in tuning and buffering gene expressions in a number of critical cellular processes, have long attracted evolutionary biologists. However, genome-wide perspectives on their origins, potential mechanisms of their de novo generation and subsequent evolution remain largely unsolved in flowering plants. Here, genome-wide analyses of Oryza sativa and Arabidopsis thaliana revealed apparently divergent patterns of miRNA gene origins. A large proportion of miRNA genes in O. sativa were TE-related and MITE-related miRNAs in particular, whereas the fraction of these miRNA genes much decreased in A. thaliana. Our results show that the majority of TE-related and pseudogene-related miRNA genes have originated through inverted duplication instead of segmental or tandem duplication events. Based on the presented findings, we hypothesize and illustrate the four likely molecular mechanisms to de novo generate novel miRNA genes from TEs and pseudogenes. Our rice genome analysis demonstrates that non-MITEs and MITEs mediated inverted duplications have played different roles in de novo generating miRNA genes. It is confirmed that the previously proposed inverted duplication model may give explanations for non-MITEs mediated duplication events. However, many other miRNA genes, known from the earlier proposed model, were rather arisen from MITE transpositions into target genes to yield binding sites. We further investigated evolutionary processes spawned from de novo generated to maturely-formed miRNA genes and their regulatory systems. We found that miRNAs increase the tunability of some gene regulatory systems with low gene copy numbers. The results also suggest that gene balance effects may have largely contributed to the evolution of miRNA regulatory systems.
Conflict of interest statement
Figures

Similar articles
-
Scenarios for the emergence of new microRNA genes in the plant Arabidopsis halleri.Plant Cell Physiol. 2025 May 17;66(4):542-553. doi: 10.1093/pcp/pcaf004. Plant Cell Physiol. 2025. PMID: 39820477
-
Dual coding of siRNAs and miRNAs by plant transposable elements.RNA. 2008 May;14(5):814-21. doi: 10.1261/rna.916708. Epub 2008 Mar 26. RNA. 2008. PMID: 18367716 Free PMC article.
-
Characterization and evolution of microRNA genes derived from repetitive elements and duplication events in plants.PLoS One. 2012;7(4):e34092. doi: 10.1371/journal.pone.0034092. Epub 2012 Apr 16. PLoS One. 2012. PMID: 22523544 Free PMC article.
-
Plant microRNAs: an insight into their gene structures and evolution.Semin Cell Dev Biol. 2010 Oct;21(8):782-9. doi: 10.1016/j.semcdb.2010.07.009. Epub 2010 Aug 4. Semin Cell Dev Biol. 2010. PMID: 20691276 Review.
-
The evolution of microRNAs in plants.Curr Opin Plant Biol. 2017 Feb;35:61-67. doi: 10.1016/j.pbi.2016.11.006. Epub 2016 Nov 22. Curr Opin Plant Biol. 2017. PMID: 27886593 Free PMC article. Review.
Cited by
-
Characterization and evolution of conserved MicroRNA through duplication events in date palm (Phoenix dactylifera).PLoS One. 2013 Aug 8;8(8):e71435. doi: 10.1371/journal.pone.0071435. eCollection 2013. PLoS One. 2013. PMID: 23951162 Free PMC article.
-
The expanding world of small RNAs in plants.Nat Rev Mol Cell Biol. 2015 Dec;16(12):727-41. doi: 10.1038/nrm4085. Epub 2015 Nov 4. Nat Rev Mol Cell Biol. 2015. PMID: 26530390 Free PMC article. Review.
-
Big effects of small RNAs on legume root biotic interactions.Genome Biol. 2014 Sep 24;15(9):475. doi: 10.1186/s13059-014-0475-2. Genome Biol. 2014. PMID: 25315222 Free PMC article.
-
PlanTE-MIR DB: a database for transposable element-related microRNAs in plant genomes.Funct Integr Genomics. 2016 May;16(3):235-42. doi: 10.1007/s10142-016-0480-5. Epub 2016 Feb 18. Funct Integr Genomics. 2016. PMID: 26887375
-
A novel Transposable element-derived microRNA participates in plant immunity to rice blast disease.Plant Biotechnol J. 2021 Sep;19(9):1798-1811. doi: 10.1111/pbi.13592. Epub 2021 Apr 9. Plant Biotechnol J. 2021. PMID: 33780108 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

