Mathematical modeling and validation of the ergosterol pathway in Saccharomyces cerevisiae
- PMID: 22194828
- PMCID: PMC3237449
- DOI: 10.1371/journal.pone.0028344
Mathematical modeling and validation of the ergosterol pathway in Saccharomyces cerevisiae
Abstract
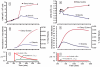
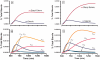
The de novo biosynthetic machinery for both sphingolipid and ergosterol production in yeast is localized in the endoplasmic reticulum (ER) and Golgi. The interconnections between the two pathways are still poorly understood, but they may be connected in specialized membrane domains, and specific knockouts strongly suggest that both routes have different layers of mutual control and are co-affected by drugs. With the goal of shedding light on the functional integration of the yeast sphingolipid-ergosterol (SL-E) pathway, we constructed a dynamic model of the ergosterol pathway using the guidelines of Biochemical Systems Theory (BST) (Savageau., J. theor. Biol., 25, 365-9, 1969). The resulting model was merged with a previous mathematical model of sphingolipid metabolism in yeast (Alvarez-Vasquez et al., J. theor. Biol., 226, 265-91, 2004; Alvarez-Vasquez et al., Nature433, 425-30, 2005). The S-system format within BST was used for analyses of consistency, stability, and sensitivity of the SL-E model, while the GMA format was used for dynamic simulations and predictions. Model validation was accomplished by comparing predictions from the model with published results on sterol and sterol-ester dynamics in yeast. The validated model was used to predict the metabolomic dynamics of the SL-E pathway after drug treatment. Specifically, we simulated the action of drugs affecting sphingolipids in the endoplasmic reticulum and studied changes in ergosterol associated with microdomains of the plasma membrane (PM).
Conflict of interest statement
Figures


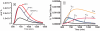

References
-
- Swain E, Baudry K, Stukey J, McDonough V, Germann M, et al. Sterol-dependent regulation of sphingolipid metabolism in Saccharomyces cerevisiae. J Biol Chem. 2002;277:26177–26184. - PubMed
-
- Fantini J, Garmy N, Mahfoud R, Yahi N. Lipid rafts: structure, function and role in HIV, Alzheimer's and prion diseases. Expert Rev Mol Med. 2002;4:1–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

