Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- PMID: 22194834
- PMCID: PMC3237453
- DOI: 10.1371/journal.pone.0028425
Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry
Abstract
Background: MALDI-TOF MS recently emerged as a valuable identification tool for bacteria and yeasts and revolutionized the daily clinical laboratory routine. But it has not been established for routine mould identification. This study aimed to validate a standardized procedure for MALDI-TOF MS-based mould identification in clinical laboratory.
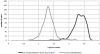
Materials and methods: First, pre-extraction and extraction procedures were optimized. With this standardized procedure, a 143 mould strains reference spectra library was built. Then, the mould isolates cultured from sequential clinical samples were prospectively subjected to this MALDI-TOF MS based-identification assay. MALDI-TOF MS-based identification was considered correct if it was concordant with the phenotypic identification; otherwise, the gold standard was DNA sequence comparison-based identification.
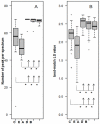
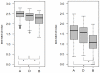
Results: The optimized procedure comprised a culture on sabouraud-gentamicin-chloramphenicol agar followed by a chemical extraction of the fungal colonies with formic acid and acetonitril. The identification was done using a reference database built with references from at least four culture replicates. For five months, 197 clinical isolates were analyzed; 20 were excluded because they were not identified at the species level. MALDI-TOF MS-based approach correctly identified 87% (154/177) of the isolates analyzed in a routine clinical laboratory activity. It failed in 12% (21/177), whose species were not represented in the reference library. MALDI-TOF MS-based identification was correct in 154 out of the remaining 156 isolates. One Beauveria bassiana was not identified and one Rhizopus oryzae was misidentified as Mucor circinelloides.
Conclusions: This work's seminal finding is that a standardized procedure can also be used for MALDI-TOF MS-based identification of a wide array of clinically relevant mould species. It thus makes it possible to identify moulds in the routine clinical laboratory setting and opens new avenues for the development of an integrated MALDI-TOF MS-based solution for the identification of any clinically relevant microorganism.
Conflict of interest statement
Figures


References
-
- Chabasse B. Emergence of new fungal pathogens: general review. Revue Francophone des Laboratoire. 2009;416:71–87.
-
- Tang CM, Holden DW, Aufauvre-Brown A, Cohen J. The detection of Aspergillus spp. by the polymerase chain reaction and its evaluation in bronchoalveolar lavage fluid. Am Rev Respir Dis. 1993;148:1313–1317. - PubMed
-
- Alexander BD. Diagnosis of fungal infection: new technologies for the mycology laboratory. Transpl Infect DisSuppl. 2002;3:32–37. - PubMed
-
- Balajee SA, Sigler L, Brandt ME. DNA and the classical way: identification of medically important molds in the 21st century. Med Mycol. 2007;45:475–490. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

