High-resolution melting genotyping of Enterococcus faecium based on multilocus sequence typing derived single nucleotide polymorphisms
- PMID: 22195020
- PMCID: PMC3241712
- DOI: 10.1371/journal.pone.0029189
High-resolution melting genotyping of Enterococcus faecium based on multilocus sequence typing derived single nucleotide polymorphisms
Abstract
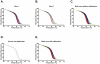
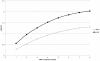
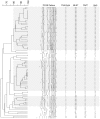
We have developed a single nucleotide polymorphism (SNP) nucleated high-resolution melting (HRM) technique to genotype Enterococcus faecium. Eight SNPs were derived from the E. faecium multilocus sequence typing (MLST) database and amplified fragments containing these SNPs were interrogated by HRM. We tested the HRM genotyping scheme on 85 E. faecium bloodstream isolates and compared the results with MLST, pulsed-field gel electrophoresis (PFGE) and an allele specific real-time PCR (AS kinetic PCR) SNP typing method. In silico analysis based on predicted HRM curves according to the G+C content of each fragment for all 567 sequence types (STs) in the MLST database together with empiric data from the 85 isolates demonstrated that HRM analysis resolves E. faecium into 231 "melting types" (MelTs) and provides a Simpson's Index of Diversity (D) of 0.991 with respect to MLST. This is a significant improvement on the AS kinetic PCR SNP typing scheme that resolves 61 SNP types with D of 0.95. The MelTs were concordant with the known ST of the isolates. For the 85 isolates, there were 13 PFGE patterns, 17 STs, 14 MelTs and eight SNP types. There was excellent concordance between PFGE, MLST and MelTs with Adjusted Rand Indices of PFGE to MelT 0.936 and ST to MelT 0.973. In conclusion, this HRM based method appears rapid and reproducible. The results are concordant with MLST and the MLST based population structure.
Conflict of interest statement
Figures


References
-
- Willems RJ, van Schaik W. Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol. 2009;4:1125–1135. - PubMed
-
- Price EP, Thiruvenkataswamy V, Mickan L, Unicomb L, Rios RE, et al. Genotyping of Campylobacter jejuni using seven single-nucleotide polymorphisms in combination with flaA short variable region sequencing. J Med Microbiol. 2006;55:1061–1070. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

