A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis
- PMID: 22195556
- PMCID: PMC3407843
- DOI: 10.1016/j.chembiol.2011.10.013
A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis
Abstract
The lipidic envelope of Mycobacterium tuberculosis promotes virulence in many ways, so we developed a lipidomics platform for a broad survey of cell walls. Here we report two new databases (MycoMass, MycoMap), 30 lipid fine maps, and mass spectrometry datasets that comprise a static lipidome. Further, by rapidly regenerating lipidomic datasets during biological processes, comparative lipidomics provides statistically valid, organism-wide comparisons that broadly assess lipid changes during infection or among clinical strains of mycobacteria. Using stringent data filters, we tracked more than 5,000 molecular features in parallel with few or no false-positive molecular discoveries. The low error rates allowed chemotaxonomic analyses of mycobacteria, which describe the extent of chemical change in each strain and identified particular strain-specific molecules for use as biomarkers.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures


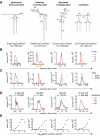
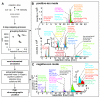


Comment in
-
TB lipidomics--the final frontier.Chem Biol. 2011 Dec 23;18(12):1517-8. doi: 10.1016/j.chembiol.2011.12.003. Chem Biol. 2011. PMID: 22195552 Free PMC article.
References
-
- Benjamini Y, Hocheberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300.
-
- Borgstrom B. Investigation on lipid separation methods. Separation of phospholipids from neutral fat and fatty acids. Acta Physiol Scand. 1952;25:101–110. - PubMed
-
- Casas-Arce E, B. t.H., B.L. F, A.J. M. Asymmetric total synthesis of PDIM A: a virulence factor of Mycobacterium tuberculosis. Chemistry. 2008;14:4157–4159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

