The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation
- PMID: 22195744
- PMCID: PMC3248798
- DOI: 10.1016/j.immuni.2011.09.021
The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation
Abstract
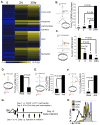
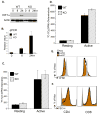
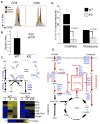
To fulfill the bioenergetic and biosynthetic demand of proliferation, T cells reprogram their metabolic pathways from fatty acid β-oxidation and pyruvate oxidation via the TCA cycle to the glycolytic, pentose-phosphate, and glutaminolytic pathways. Two of the top-ranked candidate transcription factors potentially responsible for the activation-induced T cell metabolic transcriptome, HIF1α and Myc, were induced upon T cell activation, but only the acute deletion of Myc markedly inhibited activation-induced glycolysis and glutaminolysis in T cells. Glutamine deprivation compromised activation-induced T cell growth and proliferation, and this was partially replaced by nucleotides and polyamines, implicating glutamine as an important source for biosynthetic precursors in active T cells. Metabolic tracer analysis revealed a Myc-dependent metabolic pathway linking glutaminolysis to the biosynthesis of polyamines. Therefore, a Myc-dependent global metabolic transcriptome drives metabolic reprogramming in activated, primary T lymphocytes. This may represent a general mechanism for metabolic reprogramming under patho-physiological conditions.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




Comment in
-
T cell Myc-tabolism.Immunity. 2011 Dec 23;35(6):845-6. doi: 10.1016/j.immuni.2011.12.001. Immunity. 2011. PMID: 22195738 Free PMC article.
References
-
- Bowlin TL, McKown BJ, Babcock GF, Sunkara PS. Intracellular polyamine biosynthesis is required for interleukin 2 responsiveness during lymphocyte mitogenesis. Cell Immunol. 1987;106:420–427. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

