Genomic imprinting: recognition and marking of imprinted loci
- PMID: 22195775
- PMCID: PMC3314145
- DOI: 10.1016/j.gde.2011.12.001
Genomic imprinting: recognition and marking of imprinted loci
Abstract
Genomic imprinting is an epigenetic process resulting in the monoallelic parent-of-origin-specific expression of a subset of genes in the mammalian genome. The parental alleles are differentially marked by DNA methylation during gametogenesis when the genomes are in separate compartments. How methylation machinery recognizes and differentially modifies these imprinted regions in germ cells remains a key question in the field. While studies have focused on determining a sequence signature that alone could distinguish imprinted regions from the rest of the genome, recent reports do not support such a hypothesis. Rather, it is becoming clear that features such as transcription, histone modifications and higher order chromatin are employed either individually or in combination to set up parental imprints.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
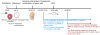

References
-
- Barlow DP. Genomic Imprinting: A Mammalian Epigenetic Discovery Model. Annu Rev Genet. 2011;45:379–404. - PubMed
-
- Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565–575. - PubMed
-
- Tomizawa Si, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138:811–820. 15 germline DMRs were examined in mouse sperm, oocytes and embryos and it was determined that maternal gametic DMRs appeared as unmethylated islands in male germ cells, the extent of gametic DMRs differs significantly in germ cells compared to embryos and substantial non-CpG DNA methylation was evident in oocytes. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

