Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells
- PMID: 22196736
- PMCID: PMC3312319
- DOI: 10.1016/j.cell.2011.09.057
Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells
Abstract
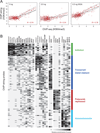
Hundreds of chromatin regulators (CRs) control chromatin structure and function by catalyzing and binding histone modifications, yet the rules governing these key processes remain obscure. Here, we present a systematic approach to infer CR function. We developed ChIP-string, a meso-scale assay that combines chromatin immunoprecipitation with a signature readout of 487 representative loci. We applied ChIP-string to screen 145 antibodies, thereby identifying effective reagents, which we used to map the genome-wide binding of 29 CRs in two cell types. We found that specific combinations of CRs colocalize in characteristic patterns at distinct chromatin environments, at genes of coherent functions, and at distal regulatory elements. When comparing between cell types, CRs redistribute to different loci but maintain their modular and combinatorial associations. Our work provides a multiplex method that substantially enhances the ability to monitor CR binding, presents a large resource of CR maps, and reveals common principles for combinatorial CR function.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
-
- Elsasser SJ, Allis CD, Lewis PW. Cancer. New epigenetic drivers of cancers. Science. 2011;331:1145–1146. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

