B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen
- PMID: 22197976
- PMCID: PMC3262910
- DOI: 10.1038/ni.2194
B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen
Erratum in
- Nat Immunol. 2014 Feb;15(2):205
Abstract
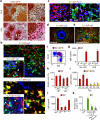
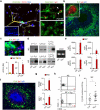
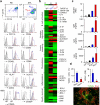
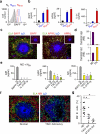
Neutrophils use immunoglobulins to clear antigen, but their role in immunoglobulin production is unknown. Here we identified neutrophils around the marginal zone (MZ) of the spleen, a B cell area specialized in T cell-independent immunoglobulin responses to circulating antigen. Neutrophils colonized peri-MZ areas after postnatal mucosal colonization by microbes and enhanced their B cell-helper function after receiving reprogramming signals, including interleukin 10 (IL-10), from splenic sinusoidal endothelial cells. Splenic neutrophils induced immunoglobulin class switching, somatic hypermutation and antibody production by activating MZ B cells through a mechanism that involved the cytokines BAFF, APRIL and IL-21. Neutropenic patients had fewer and hypomutated MZ B cells and a lower abundance of preimmune immunoglobulins to T cell-independent antigens, which indicates that neutrophils generate an innate layer of antimicrobial immunoglobulin defense by interacting with MZ B cells.
Figures

Comment in
-
A helping hand from neutrophils in T cell-independent antibody responses?Nat Immunol. 2012 Jan 19;13(2):111-3. doi: 10.1038/ni.2214. Nat Immunol. 2012. PMID: 22261958 No abstract available.
-
Antibody responses: Neutrophils zone in to help B cells.Nat Rev Immunol. 2012 Jan 20;12(2):73. doi: 10.1038/nri3159. Nat Rev Immunol. 2012. PMID: 22266690 No abstract available.
References
-
- Nathan C. Neutrophils and immunity: challenges and opportunities. Nat. Rev. Immunol. 2006;6:173–182. - PubMed
-
- Soehnlein O. An elegant defense: how neutrophils shape the immune response. Trends Immunol. 2009;30:511–512. - PubMed
-
- Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. - PubMed
-
- Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011;11:519–531. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P51 RR00165/RR/NCRR NIH HHS/United States
- P01 096187/PHS HHS/United States
- U01 AI095613/AI/NIAID NIH HHS/United States
- P51 RR000165/RR/NCRR NIH HHS/United States
- P01 AI061093/AI/NIAID NIH HHS/United States
- U01 AI95613/AI/NIAID NIH HHS/United States
- R01 AI074378/AI/NIAID NIH HHS/United States
- P01 AI096187/AI/NIAID NIH HHS/United States
- P01 DK072201/DK/NIDDK NIH HHS/United States
- U19 AI096187/AI/NIAID NIH HHS/United States
- P01 AI61093/AI/NIAID NIH HHS/United States
- R01 AI057653/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

