Mutability and mutational spectrum of chromosome transmission fidelity genes
- PMID: 22198145
- PMCID: PMC3350768
- DOI: 10.1007/s00412-011-0356-3
Mutability and mutational spectrum of chromosome transmission fidelity genes
Abstract
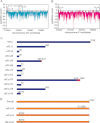
It has been more than two decades since the original chromosome transmission fidelity (Ctf) screen of Saccharomyces cerevisiae was published. Since that time the spectrum of mutations known to cause Ctf and, more generally, chromosome instability (CIN) has expanded dramatically as a result of systematic screens across yeast mutant arrays. Here we describe a comprehensive summary of the original Ctf genetic screen and the cloning of the remaining complementation groups as efforts to expand our knowledge of the CIN gene repertoire and its mutability in a model eukaryote. At the time of the original screen, it was impossible to predict either the genes and processes that would be overrepresented in a pool of random mutants displaying a Ctf phenotype or what the entire set of genes potentially mutable to Ctf would be. We show that in a collection of 136 randomly selected Ctf mutants, >65% of mutants map to 13 genes, 12 of which are involved in sister chromatid cohesion and/or kinetochore function. Extensive screening of systematic mutant collections has shown that ~350 genes with functions as diverse as RNA processing and proteasomal activity mutate to cause a Ctf phenotype and at least 692 genes are required for faithful chromosome segregation. The enrichment of random Ctf alleles in only 13 of ~350 possible Ctf genes suggests that these genes are more easily mutable to cause genome instability than the others. These observations inform our understanding of recurring CIN mutations in human cancers where presumably random mutations are responsible for initiating the frequently observed CIN phenotype of tumors.
Figures


Similar articles
-
Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation.Proc Natl Acad Sci U S A. 2005 Sep 27;102(39):13956-61. doi: 10.1073/pnas.0503504102. Epub 2005 Sep 19. Proc Natl Acad Sci U S A. 2005. PMID: 16172405 Free PMC article.
-
Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae.Mol Cell Biol. 1996 Jun;16(6):2838-47. doi: 10.1128/MCB.16.6.2838. Mol Cell Biol. 1996. PMID: 8649393 Free PMC article.
-
Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation.Genetics. 2001 Apr;157(4):1543-53. doi: 10.1093/genetics/157.4.1543. Genetics. 2001. PMID: 11290711 Free PMC article.
-
Mechanisms of chromosomal instability.Curr Biol. 2010 Mar 23;20(6):R285-95. doi: 10.1016/j.cub.2010.01.034. Curr Biol. 2010. PMID: 20334839 Free PMC article. Review.
-
Studies of meiosis disclose distinct roles of cohesion in the core centromere and pericentromeric regions.Chromosome Res. 2009;17(2):239-49. doi: 10.1007/s10577-008-9013-y. Chromosome Res. 2009. PMID: 19308704 Review.
Cited by
-
Biogenesis of RNA polymerases II and III requires the conserved GPN small GTPases in Saccharomyces cerevisiae.Genetics. 2013 Mar;193(3):853-64. doi: 10.1534/genetics.112.148726. Epub 2012 Dec 24. Genetics. 2013. PMID: 23267056 Free PMC article.
-
Saccharomyces cerevisiae genetics predicts candidate therapeutic genetic interactions at the mammalian replication fork.G3 (Bethesda). 2013 Feb;3(2):273-82. doi: 10.1534/g3.112.004754. Epub 2013 Feb 1. G3 (Bethesda). 2013. PMID: 23390603 Free PMC article.
-
The sister chromatid cohesion pathway suppresses multiple chromosome gain and chromosome amplification.Genetics. 2014 Feb;196(2):373-84. doi: 10.1534/genetics.113.159202. Epub 2013 Dec 2. Genetics. 2014. PMID: 24298060 Free PMC article.
-
Exonuclease domain mutants of yeast DIS3 display genome instability.Nucleus. 2019 Dec;10(1):21-32. doi: 10.1080/19491034.2019.1578600. Nucleus. 2019. PMID: 30724665 Free PMC article.
-
Synthetic lethality and cancer: cohesin and PARP at the replication fork.Trends Genet. 2013 May;29(5):290-7. doi: 10.1016/j.tig.2012.12.004. Epub 2013 Jan 18. Trends Genet. 2013. PMID: 23333522 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

