PPARα activation inhibits endothelin-1-induced cardiomyocyte hypertrophy by prevention of NFATc4 binding to GATA-4
- PMID: 22198280
- PMCID: PMC3506254
- DOI: 10.1016/j.abb.2011.11.024
PPARα activation inhibits endothelin-1-induced cardiomyocyte hypertrophy by prevention of NFATc4 binding to GATA-4
Abstract
Peroxisome proliferator-activated receptor alpha (PPARα) has been implicated in the pathogenesis of cardiac hypertrophy, although its mechanism of action remains largely unknown. To determine the effect of PPARα activation on endothelin-1 (ET-1)-induced cardiomyocyte hypertrophy and explore its molecular mechanisms, we evaluated the interaction of PPARα with nuclear factor of activated T-cells c4 (NFATc4) in nuclei of cardiomyocytes from neonatal rats in primary culture. In ET-1-stimulated cardiomyocytes, data from electrophoretic mobility-shift assays (EMSA) and co-immunoprecipitation (co-IP) revealed that fenofibrate (Fen), a PPARα activator, in a concentration-dependent manner, enhanced the association of NFATc4 with PPARα and decreased its interaction with GATA-4, in promoter complexes involved in activation of the rat brain natriuretic peptide (rBNP) gene. Effects of PPARα overexpression were similar to those of its activation by Fen. PPARα depletion by small interfering RNA abolished inhibitory effects of Fen on NFATc4 binding to GATA-4 and the rBNP DNA. Quantitative RT-PCR and confocal microscopy confirmed inhibitory effects of PPARα activation on elevation of rBNP mRNA levels and ET-1-induced cardiomyocyte hypertrophy. Our results suggest that activated PPARα can compete with GATA-4 binding to NFATc4, thereby decreasing transactivation of NFATc4, and interfering with ET-1 induced cardiomyocyte hypertrophy.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
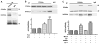


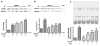
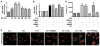
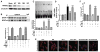

References
-
- Lloyd-Jones DM, Larson MG, Leip EP, Beiser A, D’Agostino RB, Kannel WB, Murabito JM, Vasan RS, Benjamin EJ, Levy D. Circulation. 2002;106:3068–3072. - PubMed
-
- Ho KK, Pinsky JL, Kannel WB, Levy D. Journal of the American College of Cardiology. 1993;22:6A–13A. - PubMed
-
- Finckenberg P, Mervaala E. Journal of Hypertension. 2010;28(Suppl 1):S33–38. - PubMed
-
- Eder P, Molkentin JD. Circulation Research. 2011;108:265–272. - PubMed
-
- Chien KR, Knowlton KU, Zhu H, Chien S. FASEB Journal. 1991;5:3037–3046. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

