Molecular mechanisms of EGF signaling-dependent regulation of pipe, a gene crucial for dorsoventral axis formation in Drosophila
- PMID: 22198544
- PMCID: PMC3291829
- DOI: 10.1007/s00427-011-0384-2
Molecular mechanisms of EGF signaling-dependent regulation of pipe, a gene crucial for dorsoventral axis formation in Drosophila
Abstract
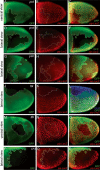
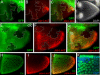
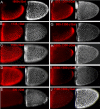
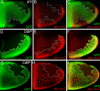
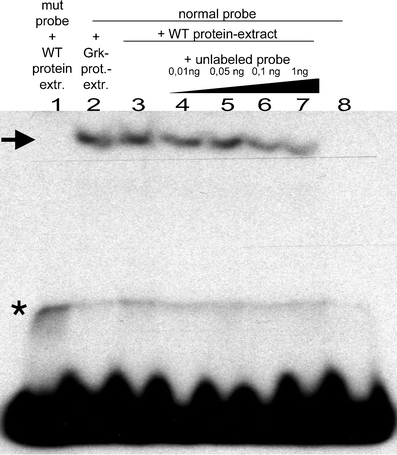
During Drosophila oogenesis the expression of the sulfotransferase Pipe in ventral follicle cells is crucial for dorsoventral axis formation. Pipe modifies proteins that are incorporated in the ventral eggshell and activate Toll signaling which in turn initiates embryonic dorsoventral patterning. Ventral pipe expression is the result of an oocyte-derived EGF signal which down-regulates pipe in dorsal follicle cells. The analysis of mutant follicle cell clones reveals that none of the transcription factors known to act downstream of EGF signaling in Drosophila is required or sufficient for pipe regulation. However, the pipe cis-regulatory region harbors a 31-bp element which is essential for pipe repression, and ovarian extracts contain a protein that binds this element. Thus, EGF signaling does not act by down-regulating an activator of pipe as previously suggested but rather by activating a repressor. Surprisingly, this repressor acts independent of the common co-repressors Groucho or CtBP.
Figures


References
-
- Atkey MR, Lachance JF, Walczak M, Rebello T, Nilson LA. Capicua regulates follicle cell fate in the Drosophila ovary through repression of mirror. Development. 2006;133:2115–2123. - PubMed
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. - PubMed
-
- Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol. 1995;3:21–29. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

