Genome architectures revealed by tethered chromosome conformation capture and population-based modeling
- PMID: 22198700
- PMCID: PMC3782096
- DOI: 10.1038/nbt.2057
Genome architectures revealed by tethered chromosome conformation capture and population-based modeling
Abstract
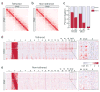
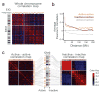
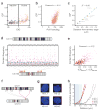
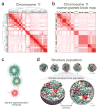
We describe tethered conformation capture (TCC), a method for genome-wide mapping of chromatin interactions. By performing ligations on solid substrates rather than in solution, TCC substantially enhances the signal-to-noise ratio, thereby facilitating a detailed analysis of interactions within and between chromosomes. We identified a group of regions in each chromosome in human cells that account for the majority of interchromosomal interactions. These regions are marked by high transcriptional activity, suggesting that their interactions are mediated by transcriptional machinery. Each of these regions interacts with numerous other such regions throughout the genome in an indiscriminate fashion, partly driven by the accessibility of the partners. As a different combination of interactions is likely present in different cells, we developed a computational method to translate the TCC data into physical chromatin contacts in a population of three-dimensional genome structures. Statistical analysis of the resulting population demonstrates that the indiscriminate properties of interchromosomal interactions are consistent with the well-known architectural features of the human genome.
Figures

Comment in
-
Parallel genome universes.Nat Biotechnol. 2012 Jan 9;30(1):55-6. doi: 10.1038/nbt.2085. Nat Biotechnol. 2012. PMID: 22231096 Free PMC article. No abstract available.
Similar articles
-
Population-based 3D genome structure analysis reveals driving forces in spatial genome organization.Proc Natl Acad Sci U S A. 2016 Mar 22;113(12):E1663-72. doi: 10.1073/pnas.1512577113. Epub 2016 Mar 7. Proc Natl Acad Sci U S A. 2016. PMID: 26951677 Free PMC article.
-
A Guided Protocol for Array Based T2C: A High-Quality Selective High-Resolution High-Throughput Chromosome Interaction Capture.Curr Protoc Hum Genet. 2018 Oct;99(1):e55. doi: 10.1002/cphg.55. Epub 2018 Sep 10. Curr Protoc Hum Genet. 2018. PMID: 30199150
-
Parallel genome universes.Nat Biotechnol. 2012 Jan 9;30(1):55-6. doi: 10.1038/nbt.2085. Nat Biotechnol. 2012. PMID: 22231096 Free PMC article. No abstract available.
-
Chromatin and epigenetic features of long-range gene regulation.Nucleic Acids Res. 2013 Aug;41(15):7185-99. doi: 10.1093/nar/gkt499. Epub 2013 Jun 13. Nucleic Acids Res. 2013. PMID: 23766291 Free PMC article. Review.
-
Simulation of different three-dimensional polymer models of interphase chromosomes compared to experiments-an evaluation and review framework of the 3D genome organization.Semin Cell Dev Biol. 2019 Jun;90:19-42. doi: 10.1016/j.semcdb.2018.07.012. Epub 2018 Aug 24. Semin Cell Dev Biol. 2019. PMID: 30125668 Review.
Cited by
-
Euchromatin islands in large heterochromatin domains are enriched for CTCF binding and differentially DNA-methylated regions.BMC Genomics. 2012 Oct 26;13:566. doi: 10.1186/1471-2164-13-566. BMC Genomics. 2012. PMID: 23102236 Free PMC article.
-
Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data.Nat Rev Genet. 2013 Jun;14(6):390-403. doi: 10.1038/nrg3454. Epub 2013 May 9. Nat Rev Genet. 2013. PMID: 23657480 Free PMC article. Review.
-
Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors.Genome Biol. 2012 Sep 26;13(9):R48. doi: 10.1186/gb-2012-13-9-r48. Genome Biol. 2012. PMID: 22950945 Free PMC article.
-
A maximum-entropy model to predict 3D structural ensembles of chromatin from pairwise distances with applications to interphase chromosomes and structural variants.Nat Commun. 2023 Mar 1;14(1):1150. doi: 10.1038/s41467-023-36412-4. Nat Commun. 2023. PMID: 36854665 Free PMC article.
-
The role of 3D genome organization in disease: From compartments to single nucleotides.Semin Cell Dev Biol. 2019 Jun;90:104-113. doi: 10.1016/j.semcdb.2018.07.005. Epub 2018 Jul 17. Semin Cell Dev Biol. 2019. PMID: 30017907 Free PMC article. Review.
References
-
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. - PubMed
-
- Branco MR, Pombo A. Chromosome organization: new facts, new models. Trends Cell Biol. 2007;17:127–134. - PubMed
-
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. - PubMed
-
- Boyle S, et al. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. - PubMed
-
- Cremer M, et al. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–567. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

