Database independent proteomics analysis of the ostrich and human proteome
- PMID: 22198768
- PMCID: PMC3258627
- DOI: 10.1073/pnas.1108399108
Database independent proteomics analysis of the ostrich and human proteome
Abstract
Mass spectrometry (MS)-based proteome analysis relies heavily on the presence of complete protein databases. Such a strategy is extremely powerful, albeit not adequate in the analysis of unpredicted postgenome events, such as posttranslational modifications, which exponentially increase the search space. Therefore, it is of interest to explore "database-free" approaches. Here, we sampled the ostrich and human proteomes with a method facilitating de novo sequencing, utilizing the protease Lys-N in combination with electron transfer dissociation. By implementing several validation steps, including the combined use of collision-induced dissociation/electron transfer dissociation data and a cross-validation with conventional database search strategies, we identified approximately 2,500 unique de novo peptide sequences from the ostrich sample with over 900 peptides generating full backbone sequence coverage. This dataset allowed the appropriate positioning of ostrich in the evolutionary tree. The described database-free sequencing approach is generically applicable and has great potential in important proteomics applications such as in the analysis of variable parts of endogenous antibodies or proteins modified by a plethora of complex posttranslational modifications.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
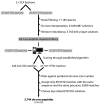


References
-
- McLafferty FW. A century of progress in molecular mass spectrometry. Annu Rev Anal Chem. 2011;4:1–22. - PubMed
-
- Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80:273–299. - PubMed
-
- Sadygov RG, Cociorva D, Yates JR., 3rd Large-scale database searching using tandem mass spectra: Looking up the answer in the back of the book. Nat Methods. 2004;1:195–202. - PubMed
-
- Pappin DJ, Hojrup P, Bleasby AJ. Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol. 1993;3:327–332. - PubMed
-
- Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–3266.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

