Gene-environment interactions in genome-wide association studies: a comparative study of tests applied to empirical studies of type 2 diabetes
- PMID: 22199026
- PMCID: PMC3261439
- DOI: 10.1093/aje/kwr368
Gene-environment interactions in genome-wide association studies: a comparative study of tests applied to empirical studies of type 2 diabetes
Abstract
The question of which statistical approach is the most effective for investigating gene-environment (G-E) interactions in the context of genome-wide association studies (GWAS) remains unresolved. By using 2 case-control GWAS (the Nurses' Health Study, 1976-2006, and the Health Professionals Follow-up Study, 1986-2006) of type 2 diabetes, the authors compared 5 tests for interactions: standard logistic regression-based case-control; case-only; semiparametric maximum-likelihood estimation of an empirical-Bayes shrinkage estimator; and 2-stage tests. The authors also compared 2 joint tests of genetic main effects and G-E interaction. Elevated body mass index was the exposure of interest and was modeled as a binary trait to avoid an inflated type I error rate that the authors observed when the main effect of continuous body mass index was misspecified. Although both the case-only and the semiparametric maximum-likelihood estimation approaches assume that the tested markers are independent of exposure in the general population, the authors did not observe any evidence of inflated type I error for these tests in their studies with 2,199 cases and 3,044 controls. Both joint tests detected markers with known marginal effects. Loci with the most significant G-E interactions using the standard, empirical-Bayes, and 2-stage tests were strongly correlated with the exposure among controls. Study findings suggest that methods exploiting G-E independence can be efficient and valid options for investigating G-E interactions in GWAS.
Figures
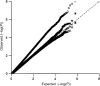
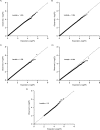
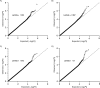
Comment in
-
Invited commentary: GE-Whiz! Ratcheting gene-environment studies up to the whole genome and the whole exposome.Am J Epidemiol. 2012 Feb 1;175(3):203-7; discussion 208-9. doi: 10.1093/aje/kwr365. Epub 2011 Dec 22. Am J Epidemiol. 2012. PMID: 22199029 Free PMC article.
References
-
- Dempfle A, Scherag A, Hein R, et al. Gene-environment interactions for complex traits: definitions, methodological requirements and challenges. Eur J Hum Genet. 2008;16(10):1164–1172. - PubMed
-
- Hwang SJ, Beaty TH, Liang KY, et al. Minimum sample size estimation to detect gene-environment interaction in case-control designs. Am J Epidemiol. 1994;140(11):1029–1037. - PubMed
-
- Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat Med. 1994;13(2):153–162. - PubMed
-
- Chatterjee N, Carroll RJ. Semiparametric maximum likelihood estimation exploiting gene-environment independence in case-control studies. Biometrika. 2005;92(2):399–418.
-
- Satten GA, Epstein MP. Comparison of prospective and retrospective methods for haplotype inference in case-control studies. Genet Epidemiol. 2004;27(3):192–201. - PubMed
Publication types
MeSH terms
Grants and funding
- P01 CA055075/CA/NCI NIH HHS/United States
- U01 HG004415/HG/NHGRI NIH HHS/United States
- HHSN268200782096C/HG/NHGRI NIH HHS/United States
- R01 DA013423/DA/NIDA NIH HHS/United States
- U01HG004436/HG/NHGRI NIH HHS/United States
- U01 HG004735/HG/NHGRI NIH HHS/United States
- U01 HG004738/HG/NHGRI NIH HHS/United States
- CA136792/CA/NCI NIH HHS/United States
- U01HG004446/HG/NHGRI NIH HHS/United States
- CA54281/CA/NCI NIH HHS/United States
- P01CA089392/CA/NCI NIH HHS/United States
- U01HG004726/HG/NHGRI NIH HHS/United States
- U01HG004438/HG/NHGRI NIH HHS/United States
- R01 DK058845/DK/NIDDK NIH HHS/United States
- U01 HG004728/HG/NHGRI NIH HHS/United States
- U01 HG004438/HG/NHGRI NIH HHS/United States
- U10AA008401/AA/NIAAA NIH HHS/United States
- P01CA087969/CA/NCI NIH HHS/United States
- U01 HG004446/HG/NHGRI NIH HHS/United States
- U01HG004738/HG/NHGRI NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- P01CA055075/CA/NCI NIH HHS/United States
- Z01 CP010200/ImNIH/Intramural NIH HHS/United States
- U01HG004415/HG/NHGRI NIH HHS/United States
- U01 DE018993/DE/NIDCR NIH HHS/United States
- U01HG004735/HG/NHGRI NIH HHS/United States
- P01 CA089392/CA/NCI NIH HHS/United States
- CAPMC/ CIHR/Canada
- U01 HG004402/HG/NHGRI NIH HHS/United States
- U01 HG004424/HG/NHGRI NIH HHS/United States
- U01HG004728/HG/NHGRI NIH HHS/United States
- U01HG004399/HG/NHGRI NIH HHS/United States
- U01HG004402/HG/NHGRI NIH HHS/United States
- U01HG004422/HG/NHGRI NIH HHS/United States
- CA63464/CA/NCI NIH HHS/United States
- R01DA013423/DA/NIDA NIH HHS/United States
- U01 HG004729/HG/NHGRI NIH HHS/United States
- HHSN-268200782096C/PHS HHS/United States
- U01 HG004422/HG/NHGRI NIH HHS/United States
- U01HG04424/HG/NHGRI NIH HHS/United States
- R01 CA063464/CA/NCI NIH HHS/United States
- R21 ES019712/ES/NIEHS NIH HHS/United States
- U01DE018993/DE/NIDCR NIH HHS/United States
- U01HG004423/HG/NHGRI NIH HHS/United States
- R01DK058845/DK/NIDDK NIH HHS/United States
- R01 CA054281/CA/NCI NIH HHS/United States
- U01DE018903/DE/NIDCR NIH HHS/United States
- U01 CA063464/CA/NCI NIH HHS/United States
- U01HG004729/HG/NHGRI NIH HHS/United States
- U01 DE018903/DE/NIDCR NIH HHS/United States
- U01 HG004436/HG/NHGRI NIH HHS/United States
- U10 AA008401/AA/NIAAA NIH HHS/United States
- U01 HG004423/HG/NHGRI NIH HHS/United States
- RFAHG006033/PHS HHS/United States
- U01 CA136792/CA/NCI NIH HHS/United States
- R37 CA054281/CA/NCI NIH HHS/United States
- U01 HG004399/HG/NHGRI NIH HHS/United States
- U01 HG004726/HG/NHGRI NIH HHS/United States
- 1R21 ES019712-01/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

