Surface interactome in Streptococcus pyogenes
- PMID: 22199230
- PMCID: PMC3322576
- DOI: 10.1074/mcp.M111.015206
Surface interactome in Streptococcus pyogenes
Abstract
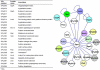
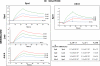
Very few studies have so far been dedicated to the systematic analysis of protein interactions occurring between surface and/or secreted proteins in bacteria. Such interactions are expected to play pivotal biological roles that deserve investigation. Taking advantage of the availability of a detailed map of surface and secreted proteins in Streptococcus pyogenes (group A Streptococcus (GAS)), we used protein array technology to define the "surface interactome" in this important human pathogen. Eighty-three proteins were spotted on glass slides in high density format, and each of the spotted proteins was probed for its capacity to interact with any of the immobilized proteins. A total of 146 interactions were identified, 25 of which classified as "reciprocal," namely, interactions that occur irrespective of which of the two partners was immobilized on the chip or in solution. Several of these interactions were validated by surface plasmon resonance and supported by confocal microscopy analysis of whole bacterial cells. By this approach, a number of interesting interactions have been discovered, including those occurring between OppA, DppA, PrsA, and TlpA, proteins known to be involved in protein folding and transport. These proteins, all localizing at the septum, might be part, together with HtrA, of the recently described ExPortal complex of GAS. Furthermore, SpeI was found to strongly interact with the metal transporters AdcA and Lmb. Because SpeI strictly requires zinc to exert its function, this finding provides evidence on how this superantigen, a major player in GAS pathogenesis, can acquire the metal in the host environment, where it is largely sequestered by carrier proteins. We believe that the approach proposed herein can lead to a deeper knowledge of the mechanisms underlying bacterial invasion, colonization, and pathogenesis.
Figures





References
-
- Nakai K. (2000) Protein sorting signals and prediction of subcellular localization. Adv. Protein Chem. 54, 277–344 - PubMed
-
- Pizza M., Scarlato V., Masignani V., Giuliani M. M., Aricò B., Comanducci M., Jennings G. T., Baldi L., Bartolini E., Capecchi B., Galeotti C. L., Luzzi E., Manetti R., Marchetti E., Mora M., Nuti S., Ratti G., Santini L., Savino S., Scarselli M., Storni E., Zuo P., Broeker M., Hundt E., Knapp B., Blair E., Mason T., Tettelin H., Hood D. W., Jeffries A. C., Saunders N. J., Granoff D. M., Venter J. C., Moxon E. R., Grandi G., Rappuoli R. (2000) Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287, 1816–1820 - PubMed
-
- Maione D., Margarit I., Rinaudo C. D., Masignani V., Mora M., Scarselli M., Tettelin H., Brettoni C., Iacobini E. T., Rosini R., D'Agostino N., Miorin L., Buccato S., Mariani M., Galli G., Nogarotto R., Nardi Dei V., Vegni F., Fraser C., Mancuso G., Teti G., Madoff L. C., Paoletti L. C., Rappuoli R., Kasper D. L., Telford J. L., Grandi G. (2005) Identification of a universal Group B Streptococcus vaccine by multiple genome screen. Science 309, 148–150 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

