Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling
- PMID: 22199352
- PMCID: PMC3285328
- DOI: 10.1074/jbc.M111.312280
Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling
Abstract
In eukaryotic cells, the spatial regulation of protein expression is frequently conferred through the coupling of mRNA localization and the local control of translation. mRNA localization to the endoplasmic reticulum (ER) is a prominent example of such regulation and serves a ubiquitous role in segregating the synthesis of secretory and integral membrane proteins to the ER. Recent genomic and biochemical studies have now expanded this view to suggest a more substantial role for the ER cellular protein synthesis. We have utilized cell fractionation and ribosome profiling to obtain a genomic survey of the subcellular organization of mRNA translation and report that ribosomal loading of mRNAs, a proxy for mRNA translation, is biased to the ER. Notably, ER-associated mRNAs encoding both cytosolic and topogenic signal-encoding proteins display similar ribosome loading densities, suggesting that ER-associated ribosomes serve a global role in mRNA translation. We propose that the partitioning of mRNAs and their translation between the cytosol and ER compartments may represent a novel mechanism for the post-transcriptional regulation of gene expression.
Figures

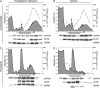
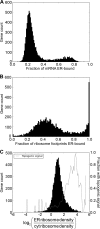
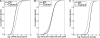
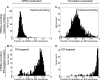

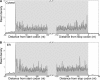
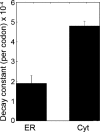
References
-
- Redman C. M. (1969) Biosynthesis of serum proteins and ferritin by free and attached ribosomes of rat liver. J. Biol. Chem. 244, 4308–4315 - PubMed
-
- Hicks S. J., Drysdale J. W., Munro H. N. (1969) Preferential synthesis of ferritin and albumin by different populations of liver polysomes. Science 164, 584–585 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

