Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data
- PMID: 22199388
- PMCID: PMC3278759
- DOI: 10.1093/bioinformatics/btr703
Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data
Abstract
Motivation: High-throughput sequencing (HTS) technologies have made low-cost sequencing of large numbers of samples commonplace. An explosion in the type, not just number, of sequencing experiments has also taken place including genome re-sequencing, population-scale variation detection, whole transcriptome sequencing and genome-wide analysis of protein-bound nucleic acids.
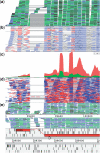
Results: We present Artemis as a tool for integrated visualization and computational analysis of different types of HTS datasets in the context of a reference genome and its corresponding annotation.
Availability: Artemis is freely available (under a GPL licence) for download (for MacOSX, UNIX and Windows) at the Wellcome Trust Sanger Institute websites: http://www.sanger.ac.uk/resources/software/artemis/.
Figures



References
-
- Carver T.J., et al. ACT: the Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

