Characterization and inhibition of norovirus proteases of genogroups I and II using a fluorescence resonance energy transfer assay
- PMID: 22200497
- PMCID: PMC3259199
- DOI: 10.1016/j.virol.2011.12.002
Characterization and inhibition of norovirus proteases of genogroups I and II using a fluorescence resonance energy transfer assay
Abstract
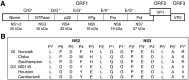
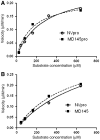
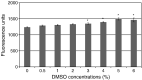
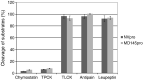
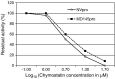
Noroviruses are the major cause of food- or water-borne gastroenteritis outbreaks in humans. The norovirus protease that cleaves a large viral polyprotein to nonstructural proteins is essential for virus replication and an attractive target for antiviral drug development. Noroviruses show high genetic diversity with at least five genogroups, GI-GV, of which GI and GII are responsible for the majority of norovirus infections in humans. We cloned and expressed proteases of Norwalk virus (GI) and MD145 virus (GII) and characterized the enzymatic activities with fluorescence resonance energy transfer substrates. We demonstrated that the GI and GII proteases cleaved the substrates derived from the naturally occurring cleavage site in the open reading frame (ORF) 1 of G1 norovirus with similar efficiency, and that enzymatic activity of both proteases was inhibited by commercial protease inhibitors including chymostatin. The interaction of chymostatin to Norwalk virus protease was validated by nuclear magnetic resonance (NMR) spectroscopy.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Expression, Purification and Characterization of a GII.4 Norovirus Protease from Minerva Virus.Infect Disord Drug Targets. 2018;18(3):224-232. doi: 10.2174/1871526518666180521091158. Infect Disord Drug Targets. 2018. PMID: 29779487 Free PMC article.
-
Development of a Gaussia luciferase-based human norovirus protease reporter system: cell type-specific profile of Norwalk virus protease precursors and evaluation of inhibitors.J Virol. 2014 Sep;88(18):10312-26. doi: 10.1128/JVI.01111-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008934 Free PMC article.
-
A Cell-based Fluorescence Resonance Energy Transfer (FRET) Sensor Reveals Inter- and Intragenogroup Variations in Norovirus Protease Activity and Polyprotein Cleavage.J Biol Chem. 2015 Nov 13;290(46):27841-53. doi: 10.1074/jbc.M115.688234. Epub 2015 Sep 11. J Biol Chem. 2015. PMID: 26363064 Free PMC article.
-
Norovirus Protease Structure and Antivirals Development.Viruses. 2021 Oct 14;13(10):2069. doi: 10.3390/v13102069. Viruses. 2021. PMID: 34696498 Free PMC article. Review.
-
[Noroviruses--tactic of spread].Przegl Epidemiol. 2009;63(1):5-9. Przegl Epidemiol. 2009. PMID: 19522218 Review. Polish.
Cited by
-
Structural and dynamics characterization of norovirus protease.Protein Sci. 2013 Mar;22(3):347-57. doi: 10.1002/pro.2215. Epub 2013 Jan 27. Protein Sci. 2013. PMID: 23319456 Free PMC article.
-
Substrate specificity of Tulane virus protease.Virology. 2013 Feb 5;436(1):24-32. doi: 10.1016/j.virol.2012.10.010. Epub 2012 Nov 8. Virology. 2013. PMID: 23141588 Free PMC article.
-
Antiviral Drug Discovery: Norovirus Proteases and Development of Inhibitors.Viruses. 2019 Feb 25;11(2):197. doi: 10.3390/v11020197. Viruses. 2019. PMID: 30823509 Free PMC article. Review.
-
Design, synthesis, and bioevaluation of viral 3C and 3C-like protease inhibitors.Bioorg Med Chem Lett. 2013 Dec 1;23(23):6317-20. doi: 10.1016/j.bmcl.2013.09.070. Epub 2013 Sep 30. Bioorg Med Chem Lett. 2013. PMID: 24125888 Free PMC article.
-
Potential Therapeutic Agents for Feline Calicivirus Infection.Viruses. 2018 Aug 16;10(8):433. doi: 10.3390/v10080433. Viruses. 2018. PMID: 30115859 Free PMC article.
References
-
- Atmar R.L., Estes M.K. The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. North Am. 2006;35(2):275–290. (viii) - PubMed
-
- Belliot G., Sosnovtsev S.V., Mitra T., Hammer C., Garfield M., Green K.Y. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J. Virol. 2003;77(20):10957–10974. - PMC - PubMed
-
- Bode W., Huber R. Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem. 1992;204(2):433–451. - PubMed
-
- C.D.C. 2010. Morbidity and Mortality Weekly Report (MMWR): Surveillance for Foodborne Disease Outbreaks — United States, 2007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

