LTR retrotransposons contribute to genomic gigantism in plethodontid salamanders
- PMID: 22200636
- PMCID: PMC3318908
- DOI: 10.1093/gbe/evr139
LTR retrotransposons contribute to genomic gigantism in plethodontid salamanders
Abstract
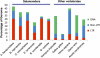
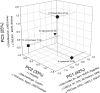
Among vertebrates, most of the largest genomes are found within the salamanders, a clade of amphibians that includes 613 species. Salamander genome sizes range from ~14 to ~120 Gb. Because genome size is correlated with nucleus and cell sizes, as well as other traits, morphological evolution in salamanders has been profoundly affected by genomic gigantism. However, the molecular mechanisms driving genomic expansion in this clade remain largely unknown. Here, we present the first comparative analysis of transposable element (TE) content in salamanders. Using high-throughput sequencing, we generated genomic shotgun data for six species from the Plethodontidae, the largest family of salamanders. We then developed a pipeline to mine TE sequences from shotgun data in taxa with limited genomic resources, such as salamanders. Our summaries of overall TE abundance and diversity for each species demonstrate that TEs make up a substantial portion of salamander genomes, and that all of the major known types of TEs are represented in salamanders. The most abundant TE superfamilies found in the genomes of our six focal species are similar, despite substantial variation in genome size. However, our results demonstrate a major difference between salamanders and other vertebrates: salamander genomes contain much larger amounts of long terminal repeat (LTR) retrotransposons, primarily Ty3/gypsy elements. Thus, the extreme increase in genome size that occurred in salamanders was likely accompanied by a shift in TE landscape. These results suggest that increased proliferation of LTR retrotransposons was a major molecular mechanism contributing to genomic expansion in salamanders.
Figures




Similar articles
-
Hellbender genome sequences shed light on genomic expansion at the base of crown salamanders.Genome Biol Evol. 2014 Jul;6(7):1818-29. doi: 10.1093/gbe/evu143. Genome Biol Evol. 2014. PMID: 25115007 Free PMC article.
-
Low levels of LTR retrotransposon deletion by ectopic recombination in the gigantic genomes of salamanders.J Mol Evol. 2015 Feb;80(2):120-9. doi: 10.1007/s00239-014-9663-7. Epub 2015 Jan 22. J Mol Evol. 2015. PMID: 25608479
-
Slow DNA loss in the gigantic genomes of salamanders.Genome Biol Evol. 2012;4(12):1340-8. doi: 10.1093/gbe/evs103. Genome Biol Evol. 2012. PMID: 23175715 Free PMC article.
-
Co-evolution of plant LTR-retrotransposons and their host genomes.Protein Cell. 2013 Jul;4(7):493-501. doi: 10.1007/s13238-013-3037-6. Epub 2013 Jun 23. Protein Cell. 2013. PMID: 23794032 Free PMC article. Review.
-
Evolution and Diversity of Transposable Elements in Vertebrate Genomes.Genome Biol Evol. 2017 Jan 1;9(1):161-177. doi: 10.1093/gbe/evw264. Genome Biol Evol. 2017. PMID: 28158585 Free PMC article. Review.
Cited by
-
A bird-like genome from a frog: Mechanisms of genome size reduction in the ornate burrowing frog, Platyplectrum ornatum.Proc Natl Acad Sci U S A. 2021 Mar 16;118(11):e2011649118. doi: 10.1073/pnas.2011649118. Proc Natl Acad Sci U S A. 2021. PMID: 33836564 Free PMC article.
-
Virus-like attachment sites as structural landmarks of plants retrotransposons.Mob DNA. 2016 Jul 28;7:14. doi: 10.1186/s13100-016-0069-5. eCollection 2016. Mob DNA. 2016. PMID: 27471551 Free PMC article.
-
Virtual Genome Walking across the 32 Gb Ambystoma mexicanum genome; assembling gene models and intronic sequence.Sci Rep. 2018 Jan 12;8(1):618. doi: 10.1038/s41598-017-19128-6. Sci Rep. 2018. PMID: 29330416 Free PMC article.
-
Genome size and chromosome number in velvet worms (Onychophora).Genetica. 2012 Dec;140(10-12):497-504. doi: 10.1007/s10709-013-9698-5. Epub 2013 Jan 11. Genetica. 2012. PMID: 23307271
-
Recurrent Co-Option and Recombination of Cytokine and Three Finger Proteins in Multiple Reproductive Tissues Throughout Salamander Evolution.Front Cell Dev Biol. 2022 Feb 23;10:828947. doi: 10.3389/fcell.2022.828947. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35281090 Free PMC article.
References
-
- Agren JA, Wright SI. Co-evolution between transposable elements and their hosts: a major factor in genome size evolution? Chromosome Res. 2011;19:777–786. - PubMed
-
- AmphibiaWeb . AmphibiaWeb: information on amphibian biology and conservation. 2011. [Internet]. Berkeley (CA): AmphibiaWeb. [cited 2011 Sep 26]. Available from: http://amphibiaweb.org.
-
- Baldari CT, Amaldi F. DNA reassociation kinetics in relation to genome size in four amphibian species. Chromosoma. 1976;59:13–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

