Cdc6 expression represses E-cadherin transcription and activates adjacent replication origins
- PMID: 22201124
- PMCID: PMC3246883
- DOI: 10.1083/jcb.201108121
Cdc6 expression represses E-cadherin transcription and activates adjacent replication origins
Abstract
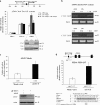
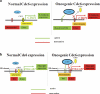
E-cadherin (CDH1) loss occurs frequently in carcinogenesis, contributing to invasion and metastasis. We observed that mouse and human epithelial cell lines overexpressing the replication licensing factor Cdc6 underwent phenotypic changes with mesenchymal features and loss of E-cadherin. Analysis in various types of human cancer revealed a strong correlation between increased Cdc6 expression and reduced E-cadherin levels. Prompted by these findings, we discovered that Cdc6 repressed CDH1 transcription by binding to the E-boxes of its promoter, leading to dissociation of the chromosomal insulator CTCF, displacement of the histone variant H2A.Z, and promoter heterochromatinization. Mutational analysis identified the Walker B motif and C-terminal region of Cdc6 as essential for CDH1 transcriptional suppression. Strikingly, CTCF displacement resulted in activation of adjacent origins of replication. These data demonstrate that Cdc6 acts as a molecular switch at the E-cadherin locus, linking transcriptional repression to activation of replication, and provide a telling example of how replication licensing factors could usurp alternative programs to fulfill distinct cellular functions.
Figures




References
-
- Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., Zoumpourlis V.C., et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 444:633–637 10.1038/nature05268 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

