Divergence hitchhiking and the spread of genomic isolation during ecological speciation-with-gene-flow
- PMID: 22201174
- PMCID: PMC3233719
- DOI: 10.1098/rstb.2011.0260
Divergence hitchhiking and the spread of genomic isolation during ecological speciation-with-gene-flow
Abstract
In allopatric populations, geographical separation simultaneously isolates the entire genome, allowing genetic divergence to accumulate virtually anywhere in the genome. In sympatric populations, however, the strong divergent selection required to overcome migration produces a genetic mosaic of divergent and non-divergent genomic regions. In some recent genome scans, each divergent genomic region has been interpreted as an independent incidence of migration/selection balance, such that the reduction of gene exchange is restricted to a few kilobases around each divergently selected gene. I propose an alternative mechanism, 'divergence hitchhiking' (DH), in which divergent selection can reduce gene exchange for several megabases around a gene under strong divergent selection. Not all genes/markers within a DH region are divergently selected, yet the entire region is protected to some degree from gene exchange, permitting genetic divergence from mechanisms other than divergent selection to accumulate secondarily. After contrasting DH and multilocus migration/selection balance (MM/SB), I outline a model in which genomic isolation at a given genomic location is jointly determined by DH and genome-wide effects of the progressive reduction in realized migration, then illustrate DH using data from several pairs of incipient species in the wild.
Figures
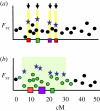
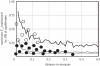

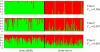
References
-
- Rice W. R., Hostert E. E. 1993. Laboratory experiments on speciation: what have we learned in 40 years? Evolution 47, 1637–1653 10.2307/2410209 (doi:10.2307/2410209) - DOI - PubMed
-
- Via S. 2001. Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol. Evol. 16, 381–390 10.1016/S0169-5347(01)02188-7 (doi:10.1016/S0169-5347(01)02188-7) - DOI - PubMed
-
- Coyne J. A., Orr H. A. 2004. Speciation. Sunderland, MA: Sinauer Associates
-
- Felsenstein J. 1981. Skepticism toward Santa Rosalia, or why are there so few kinds of animals? Evolution 35, 124–138 10.2307/2407946 (doi:10.2307/2407946) - DOI - PubMed
-
- Schluter D. 2001. Ecology and the origin of species. Trends Ecol. Evol. 16, 372–380 10.1016/S0169-5347(01)02198-X (doi:10.1016/S0169-5347(01)02198-X) - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
