Thermodynamic dissection of estrogen receptor-promoter interactions reveals that steroid receptors differentially partition their self-association and promoter binding energetics
- PMID: 22201220
- PMCID: PMC3535315
- DOI: 10.1021/bi2017156
Thermodynamic dissection of estrogen receptor-promoter interactions reveals that steroid receptors differentially partition their self-association and promoter binding energetics
Abstract
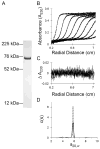
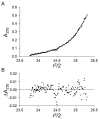
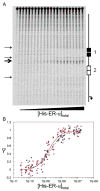
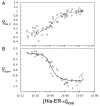
Steroid receptors define a family of ligand-activated transcription factors. Recent work has demonstrated that the receptors regulate distinct but overlapping gene networks, yet the mechanisms by which they do so remain unclear. We previously determined the microscopic binding energetics for progesterone receptor (PR) isoform assembly at promoters containing multiple response elements. We found that the two isoforms (PR-A and PR-B) share nearly identical dimerization and intrinsic DNA binding free energies but maintain large differences in cooperative free energy. Moreover, cooperativity can be modulated by monovalent ion binding and promoter layout, suggesting that differences in cooperativity might control isoform-specific promoter occupancy and thus receptor function. To determine whether cooperative binding energetics are common to other members of the steroid receptor family, we dissected the thermodynamics of estrogen receptor-α (ER-α):promoter interactions. We find that the ER-α intrinsic DNA binding free energy is identical to that of the PR isoforms. This was expected, noting that receptor DNA binding domains are highly conserved. Unexpectedly, ER-α generates negligible cooperativity-orders of magnitude less than predicted based on our studies of the PR isoforms. However, analysis of the cooperativity term suggests that it reflects a balance between highly favorable cooperative stabilization and unfavorable promoter bending. Moreover, ER-α cooperative free energy is compensated for by a large increase in dimerization free energy. Collectively, the results demonstrate that steroid receptors differentially partition not only cooperative energetics but also dimerization energetics. We speculate that this ability serves as a framework for regulating receptor-specific promoter occupancy and thus receptor-specific gene regulation.
Figures




Similar articles
-
Dissection of androgen receptor-promoter interactions: steroid receptors partition their interaction energetics in parallel with their phylogenetic divergence.J Mol Biol. 2013 Nov 15;425(22):4223-35. doi: 10.1016/j.jmb.2013.07.033. Epub 2013 Aug 3. J Mol Biol. 2013. PMID: 23917122 Free PMC article.
-
Glucocorticoid receptor-promoter interactions: energetic dissection suggests a framework for the specificity of steroid receptor-mediated gene regulation.Biochemistry. 2012 Jun 5;51(22):4463-72. doi: 10.1021/bi3003956. Epub 2012 May 22. Biochemistry. 2012. PMID: 22587663 Free PMC article.
-
Homologous steroid receptors assemble at identical promoter architectures with unique energetics of cooperativity.Proteins. 2014 Sep;82(9):2078-87. doi: 10.1002/prot.24563. Epub 2014 Apr 16. Proteins. 2014. PMID: 24648119
-
Mechanisms of action and cross-talk between estrogen receptor and progesterone receptor pathways.J Soc Gynecol Investig. 2000 Jan-Feb;7(1 Suppl):S33-7. doi: 10.1016/s1071-5576(99)00058-1. J Soc Gynecol Investig. 2000. PMID: 10732327 Review.
-
Estrogen and progesterone receptors: from molecular structures to clinical targets.Cell Mol Life Sci. 2009 Aug;66(15):2405-26. doi: 10.1007/s00018-009-0017-3. Epub 2009 Mar 31. Cell Mol Life Sci. 2009. PMID: 19333551 Free PMC article. Review.
Cited by
-
Dissection of androgen receptor-promoter interactions: steroid receptors partition their interaction energetics in parallel with their phylogenetic divergence.J Mol Biol. 2013 Nov 15;425(22):4223-35. doi: 10.1016/j.jmb.2013.07.033. Epub 2013 Aug 3. J Mol Biol. 2013. PMID: 23917122 Free PMC article.
-
Entropy tree networks of residue dynamics encode protein allostery.bioRxiv [Preprint]. 2025 Jul 1:2025.05.28.656549. doi: 10.1101/2025.05.28.656549. bioRxiv. 2025. PMID: 40501711 Free PMC article. Preprint.
-
A hydrophobic ratchet entrenches molecular complexes.Nature. 2020 Dec;588(7838):503-508. doi: 10.1038/s41586-020-3021-2. Epub 2020 Dec 9. Nature. 2020. PMID: 33299178 Free PMC article.
-
Glucocorticoid receptor-promoter interactions: energetic dissection suggests a framework for the specificity of steroid receptor-mediated gene regulation.Biochemistry. 2012 Jun 5;51(22):4463-72. doi: 10.1021/bi3003956. Epub 2012 May 22. Biochemistry. 2012. PMID: 22587663 Free PMC article.
-
Steroid receptor-DNA interactions: toward a quantitative connection between energetics and transcriptional regulation.Nucleic Acids Res. 2014 Jan;42(2):691-700. doi: 10.1093/nar/gkt859. Epub 2013 Sep 24. Nucleic Acids Res. 2014. PMID: 24064251 Free PMC article. Review.
References
-
- Tsai MJ, O’Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. - PubMed
-
- Richer JK, Jacobsen BM, Manning NG, Abel MG, Wolf DM, Horwitz KB. Differential gene regulation by the two progesterone receptor isoforms in human breast cancer cells. J Biol Chem. 2002;277:5209–5218. - PubMed
-
- Wan Y, Nordeen SK. Overlapping but distinct gene regulation profiles by glucocorticoids and progestins in human breast cancer cells. Mol Endocrinol. 2002;16:1204–1214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

