Profiling of linker histone variants in ovarian cancer
- PMID: 22201751
- PMCID: PMC3754803
- DOI: 10.2741/3934
Profiling of linker histone variants in ovarian cancer
Abstract
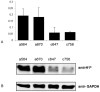
H1 linker histones play a key role in facilitating higher order chromatin folding. Emerging evidence suggests that H1 and its multiple variants are important epigenetic factors in modulating chromatin function and gene expression. Ovarian cancer is a devastating disease, ranking the fifth leading cause of all women cancer death due to its poor prognosis and difficulty in early diagnosis. Although epigenetic alterations in ovarian cancers are being appreciated in general, the role of H1 has not been explored. Here, using quantitative RT-PCR assays, we systematically examined the expression of 7 H1 genes in 33 human epithelial ovarian tumors. Whereas the expression of H1.3 was markedly increased, the expression of H10, H1.1, H1.4 and H1x were significantly reduced in malignant adenocarcinomas compared with benign adenomas. Strikingly, ovarian adenocarcinomas and adenomas exhibited characteristic expression patterns, and expression profiling of 7 H1 genes in tumor samples discriminated adenocarcinomas vs. adenomas with high accuracy. These findings indicate that the expression of H1 variants is exquisitely regulated and may serve as potential epigenetic biomarkers for ovarian cancer.
Figures



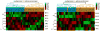
References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. - PubMed
-
- Widschwendter M, Jiang G, Woods C, Muller HM, Fiegl H, Goebel G, Marth C, Muller-Holzner E, Zeimet AG, Laird PW, Ehrlich M. DNA hypomethylation and ovarian cancer biology. Cancer Res. 2004;64:4472–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

