The natural history of ubiquitin and ubiquitin-related domains
- PMID: 22201813
- PMCID: PMC5881585
- DOI: 10.2741/3996
The natural history of ubiquitin and ubiquitin-related domains
Abstract
The ubiquitin (Ub) system is centered on conjugation and deconjugation of Ub and Ub-like (Ubls) proteins by a system of ligases and peptidases, respectively. Ub/Ubls contain the beta-grasp fold, also found in numerous proteins with biochemically distinct roles unrelated to the conventional Ub-system. The beta-GF underwent an early radiation spawning at least seven clades prior to the divergence of extant organisms from their last universal common ancestor, first emerging in the context of translation-related RNA-interactions and subsequently exploding to occupy various functional niches. Most beta-GF diversification occurred in prokaryotes, with the Ubl clade showing dramatic expansion in the eukaryotes. Diversification of Ubl families in eukaryotes played a major role in emergence of characteristic eukaryotic cellular sub-structures and systems. Recent comparative genomics studies indicate precursors of the eukaryotic Ub-system emerged in prokaryotes. The simplest of these combine an Ubl and an E1-like enzyme in metabolic pathways. Sampylation in archaea and Urmylation in eukaryotes appear to represent recruitment of such systems as simple protein-tagging apparatuses. However, other prokaryotic systems incorporated further components and mirror the eukaryotic condition in possessing an E2, a RING-type E3 or both of these components. Additionally, prokaryotes have evolved conjugation systems independent of Ub ligases, such as the Pup system.
Figures
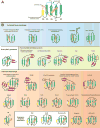

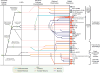


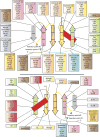
Similar articles
-
Structure and evolution of ubiquitin and ubiquitin-related domains.Methods Mol Biol. 2012;832:15-63. doi: 10.1007/978-1-61779-474-2_2. Methods Mol Biol. 2012. PMID: 22350875
-
Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system.Mol Biosyst. 2011 Jul;7(7):2261-77. doi: 10.1039/c1mb05061c. Epub 2011 May 6. Mol Biosyst. 2011. PMID: 21547297 Free PMC article.
-
Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold.Biol Direct. 2007 Jul 2;2:18. doi: 10.1186/1745-6150-2-18. Biol Direct. 2007. PMID: 17605815 Free PMC article.
-
Chemical Tools for Probing the Ub/Ubl Conjugation Cascades.Chembiochem. 2025 Jan 2;26(1):e202400659. doi: 10.1002/cbic.202400659. Epub 2024 Nov 6. Chembiochem. 2025. PMID: 39313481 Free PMC article. Review.
-
Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways.Curr Opin Struct Biol. 2007 Dec;17(6):726-35. doi: 10.1016/j.sbi.2007.08.018. Epub 2007 Oct 4. Curr Opin Struct Biol. 2007. PMID: 17919899 Free PMC article. Review.
Cited by
-
The Ubiquitin Proteasome System as a Double Agent in Plant-Virus Interactions.Plants (Basel). 2021 May 6;10(5):928. doi: 10.3390/plants10050928. Plants (Basel). 2021. PMID: 34066628 Free PMC article. Review.
-
Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling.Nucleic Acids Res. 2015 Dec 15;43(22):10633-54. doi: 10.1093/nar/gkv1267. Epub 2015 Nov 20. Nucleic Acids Res. 2015. PMID: 26590262 Free PMC article.
-
A proteomics approach to identify targets of the ubiquitin-like molecule Urm1 in Drosophila melanogaster.PLoS One. 2017 Sep 27;12(9):e0185611. doi: 10.1371/journal.pone.0185611. eCollection 2017. PLoS One. 2017. PMID: 28953965 Free PMC article.
-
Mycobacterium tuberculosis protein kinase G acts as an unusual ubiquitinating enzyme to impair host immunity.EMBO Rep. 2021 Jun 4;22(6):e52175. doi: 10.15252/embr.202052175. Epub 2021 May 2. EMBO Rep. 2021. PMID: 33938130 Free PMC article.
-
Identification of Uncharacterized Components of Prokaryotic Immune Systems and Their Diverse Eukaryotic Reformulations.J Bacteriol. 2020 Nov 19;202(24):e00365-20. doi: 10.1128/JB.00365-20. Print 2020 Nov 19. J Bacteriol. 2020. PMID: 32868406 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

