Characterization of resistance to the nonnucleoside NS5B inhibitor filibuvir in hepatitis C virus-infected patients
- PMID: 22203605
- PMCID: PMC3294886
- DOI: 10.1128/AAC.05611-11
Characterization of resistance to the nonnucleoside NS5B inhibitor filibuvir in hepatitis C virus-infected patients
Abstract
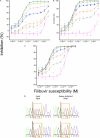
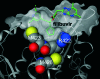
Filibuvir (PF-00868554) is an investigational nonnucleoside inhibitor of the hepatitis C virus (HCV) nonstructural 5B (NS5B) RNA-dependent RNA polymerase currently in development for treating chronic HCV infection. The aim of this study was to characterize the selection of filibuvir-resistant variants in HCV-infected individuals receiving filibuvir as short (3- to 10-day) monotherapy. We identified amino acid M423 as the primary site of mutation arising upon filibuvir dosing. Through bulk cloning of clinical NS5B sequences into a transient-replicon system, and supported by site-directed mutagenesis of the Con1 replicon, we confirmed that mutations M423I/T/V mediate phenotypic resistance. Selection in patients of an NS5B mutation at M423 was associated with a reduced replicative capacity in vitro relative to the pretherapy sequence; consistent with this, reversion to wild-type M423 was observed in the majority of patients following therapy cessation. Mutations at NS5B residues R422 and M426 were detected in a small number of patients at baseline or the end of therapy and also mediate reductions in filibuvir susceptibility, suggesting these are rare but clinically relevant alternative resistance pathways. Amino acid variants at position M423 in HCV NS5B polymerase are the preferred pathway for selection of viral resistance to filibuvir in vivo.
Figures
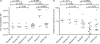
References
-
- Cooper C, et al. 2009. Evaluation of VCH-759 monotherapy in hepatitis C infection. J. Hepatol. 51:39–46 - PubMed
-
- Fried MW, et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 - PubMed
-
- Gaudieri S, et al. 2009. Hepatitis C virus drug resistance and immune-driven adaptations: relevance to new antiviral therapy. Hepatology 49:1069–1082 - PubMed
-
- Graham EJ, et al. 2011. Colony-forming assays reveal enhanced suppression of hepatitis C virus replication using combinations of direct-acting antivirals. J. Virol. Methods 174:153–157 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

