Real-Time PCR Quantification and Diversity Analysis of the Functional Genes aprA and dsrA of Sulfate-Reducing Prokaryotes in Marine Sediments of the Peru Continental Margin and the Black Sea
- PMID: 22203820
- PMCID: PMC3244613
- DOI: 10.3389/fmicb.2011.00253
Real-Time PCR Quantification and Diversity Analysis of the Functional Genes aprA and dsrA of Sulfate-Reducing Prokaryotes in Marine Sediments of the Peru Continental Margin and the Black Sea
Abstract
Sulfate-reducing prokaryotes (SRP) are ubiquitous and quantitatively important members in many ecosystems, especially in marine sediments. However their abundance and diversity in subsurface marine sediments is poorly understood. In this study, the abundance and diversity of the functional genes for the enzymes adenosine 5'-phosphosulfate reductase (aprA) and dissimilatory sulfite reductase (dsrA) of SRP in marine sediments of the Peru continental margin and the Black Sea were analyzed, including samples from the deep biosphere (ODP site 1227). For aprA quantification a Q-PCR assay was designed and evaluated. Depth profiles of the aprA and dsrA copy numbers were almost equal for all sites. Gene copy numbers decreased concomitantly with depth from around 10(8)/g sediment close to the sediment surface to less than 10(5)/g sediment at 5 mbsf. The 16S rRNA gene copy numbers of total bacteria were much higher than those of the functional genes at all sediment depths and used to calculate the proportion of SRP to the total Bacteria. The aprA and dsrA copy numbers comprised in average 0.5-1% of the 16S rRNA gene copy numbers of total bacteria in the sediments up to a depth of ca. 40 mbsf. In the zone without detectable sulfate in the pore water from about 40-121 mbsf (Peru margin ODP site 1227), only dsrA (but not aprA) was detected with copy numbers of less than 10(4)/g sediment, comprising ca. 14% of the 16S rRNA gene copy numbers of total bacteria. In this zone, sulfate might be provided for SRP by anaerobic sulfide oxidation. Clone libraries of aprA showed that all isolated sequences originate from SRP showing a close relationship to aprA of characterized species or form a new cluster with only distant relation to aprA of isolated SRP. For dsrA a high diversity was detected, even up to 121 m sediment depth in the deep biosphere.
Keywords: ODP; aprA; deep biosphere; dsrA; real-time PCR; subsurface; sulfate-reducing prokaryotes.
Figures

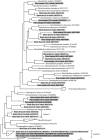
 , dsrA;
, dsrA;  , aprA.
, aprA.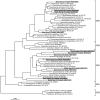
Similar articles
-
Quantification of microbial communities in near-surface and deeply buried marine sediments on the Peru continental margin using real-time PCR.Environ Microbiol. 2006 Jul;8(7):1251-60. doi: 10.1111/j.1462-2920.2006.01019.x. Environ Microbiol. 2006. PMID: 16817933
-
Quantification of Microbial Communities in Subsurface Marine Sediments of the Black Sea and off Namibia.Front Microbiol. 2012 Jan 30;3:16. doi: 10.3389/fmicb.2012.00016. eCollection 2012. Front Microbiol. 2012. PMID: 22319518 Free PMC article.
-
Abundance, diversity and activity of sulfate-reducing prokaryotes in heavy metal-contaminated sediment from a salt marsh in the Medway Estuary (UK).Mar Biotechnol (NY). 2012 Jun;14(3):363-81. doi: 10.1007/s10126-011-9420-5. Epub 2011 Nov 30. Mar Biotechnol (NY). 2012. PMID: 22124626
-
The deep biosphere in terrestrial sediments in the chesapeake bay area, virginia, USA.Front Microbiol. 2011 Jul 19;2:156. doi: 10.3389/fmicb.2011.00156. eCollection 2011. Front Microbiol. 2011. PMID: 21811489 Free PMC article.
-
Biological Sulfate Reduction in Deep Subseafloor Sediment of Guaymas Basin.Front Microbiol. 2022 Mar 3;13:845250. doi: 10.3389/fmicb.2022.845250. eCollection 2022. Front Microbiol. 2022. PMID: 35308366 Free PMC article. Review.
Cited by
-
Bacterial and archaeal communities in sediments of the north Chinese marginal seas.Microb Ecol. 2015 Jul;70(1):105-17. doi: 10.1007/s00248-014-0553-8. Epub 2014 Dec 11. Microb Ecol. 2015. PMID: 25501892
-
Impact of metabolism and growth phase on the hydrogen isotopic composition of microbial fatty acids.Front Microbiol. 2015 May 8;6:408. doi: 10.3389/fmicb.2015.00408. eCollection 2015. Front Microbiol. 2015. PMID: 26005437 Free PMC article.
-
New Dimensions in Microbial Ecology-Functional Genes in Studies to Unravel the Biodiversity and Role of Functional Microbial Groups in the Environment.Microorganisms. 2016 May 24;4(2):19. doi: 10.3390/microorganisms4020019. Microorganisms. 2016. PMID: 27681913 Free PMC article. Review.
-
Unravelling the carbon and sulphur metabolism in coastal soil ecosystems using comparative cultivation-independent genome-level characterisation of microbial communities.PLoS One. 2014 Sep 16;9(9):e107025. doi: 10.1371/journal.pone.0107025. eCollection 2014. PLoS One. 2014. PMID: 25225969 Free PMC article.
-
Phylogenetic Diversity of aprA Genes in Subseafloor Sediments on the Northwestern Pacific Margin off Japan.Microbes Environ. 2015;30(3):276-80. doi: 10.1264/jsme2.ME15023. Epub 2015 Jul 4. Microbes Environ. 2015. PMID: 26156553 Free PMC article.
References
LinkOut - more resources
Full Text Sources

