Poly(ADP-ribosyl)ation of high mobility group box 1 (HMGB1) protein enhances inhibition of efferocytosis
- PMID: 22204001
- PMCID: PMC3356430
- DOI: 10.2119/molmed.2011.00203
Poly(ADP-ribosyl)ation of high mobility group box 1 (HMGB1) protein enhances inhibition of efferocytosis
Abstract
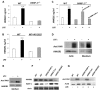
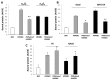
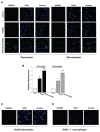
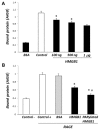
Phagocytosis of apoptotic cells by macrophages, known as efferocytosis, is a critical process in the resolution of inflammation. High mobility group box 1 (HMGB1) protein was first described as a nuclear nonhistone DNA-binding protein, but is now known to be secreted by activated cells during inflammatory processes, where it participates in diminishing efferocytosis. Although HMGB1 is known to undergo modification when secreted, the effect of such modifications on the inhibitory actions of HMGB1 during efferocytosis have not been reported. In the present studies, we found that HMGB1 secreted by Toll-like receptor 4 (TLR4) stimulated cells is highly poly(ADP-ribosyl)ated (PARylated). Gene deletion of poly(ADP)-ribose polymerase (PARP)-1 or pharmacological inhibition of PARP-1 decreased the release of HMGB1 from the nucleus to the extracellular milieu after TLR4 engagement. Preincubation of macrophages or apoptotic cells with HMGB1 diminished efferocytosis through mechanisms involving binding of HMGB1 to phosphatidylserine on apoptotic cells and to the receptor for advanced glycation end products (RAGE) on macrophages. Preincubation of either macrophages or apoptotic cells with PARylated HMGB1 inhibited efferocytosis to a greater degree than exposure to unmodified HMGB1, and PARylated HMGB1 demonstrated higher affinity for phosphatidylserine and RAGE than unmodified HMGB1. PARylated HMGB1 had a greater inhibitory effect on Ras-related C3 botulinum toxin substrate 1 (Rac-1) activation in macrophages during the uptake of apoptotic cells than unmodified HMGB1. The present results, showing that PARylation of HMGB1 enhances its ability to inhibit efferocytosis, provide a novel mechanism by which PARP-1 may promote inflammation.
Figures

Similar articles
-
PARP-1 mediates LPS-induced HMGB1 release by macrophages through regulation of HMGB1 acetylation.J Immunol. 2014 Dec 15;193(12):6114-23. doi: 10.4049/jimmunol.1400359. Epub 2014 Nov 12. J Immunol. 2014. PMID: 25392528
-
Poly(ADP-ribose) polymerase 1-sirtuin 1 functional interplay regulates LPS-mediated high mobility group box 1 secretion.Mol Med. 2015 Mar 12;20(1):612-24. doi: 10.2119/molmed.2014.00156. Mol Med. 2015. PMID: 25517228 Free PMC article.
-
HMGB1 inhibits macrophage activity in efferocytosis through binding to the alphavbeta3-integrin.Am J Physiol Cell Physiol. 2010 Dec;299(6):C1267-76. doi: 10.1152/ajpcell.00152.2010. Epub 2010 Sep 8. Am J Physiol Cell Physiol. 2010. PMID: 20826760 Free PMC article.
-
Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns.FASEB J. 2009 Mar;23(3):672-8. doi: 10.1096/fj.08-123265. Epub 2008 Nov 11. FASEB J. 2009. PMID: 19001527 Review.
-
Review: Therapeutic Targeting of HMGB1 in Stroke.Curr Drug Deliv. 2017 Sep 6;14(6):785-790. doi: 10.2174/1567201813666160808111933. Curr Drug Deliv. 2017. PMID: 27501713 Review.
Cited by
-
The Role of HMGB1 in Rheumatic Diseases.Front Immunol. 2022 Feb 17;13:815257. doi: 10.3389/fimmu.2022.815257. eCollection 2022. Front Immunol. 2022. PMID: 35250993 Free PMC article. Review.
-
Recent developments in the role of high-mobility group box 1 in systemic lupus erythematosus.Mol Med. 2014 Mar 13;20(1):72-9. doi: 10.2119/molmed.2014.00019. Mol Med. 2014. PMID: 24531837 Free PMC article. Review.
-
Molecular insights into the multifaceted functions and therapeutic targeting of high mobility group box 1 in metabolic diseases.J Cell Mol Med. 2022 Jul;26(14):3809-3815. doi: 10.1111/jcmm.17448. Epub 2022 Jun 15. J Cell Mol Med. 2022. PMID: 35706377 Free PMC article. Review.
-
PARP Inhibitors: An Innovative Approach to the Treatment of Inflammation and Metabolic Disorders in Sepsis.J Inflamm Res. 2021 May 6;14:1827-1844. doi: 10.2147/JIR.S300679. eCollection 2021. J Inflamm Res. 2021. PMID: 33986609 Free PMC article. Review.
-
Aggressiveness Niche: Can It Be the Foster Ground for Cancer Metastasis Precursors?Stem Cells Int. 2016;2016:4829106. doi: 10.1155/2016/4829106. Epub 2016 Jul 14. Stem Cells Int. 2016. PMID: 27493669 Free PMC article. Review.
References
-
- Haslett C. Granulocyte apoptosis and its role in the resolution and control of lung inflammation. Am J Respir Crit Care Med. 1999;160:S5–11. - PubMed
-
- Voll RE, et al. Immunosuppressive effects of apoptotic cells. Nature. 1997;390:350–1. - PubMed
-
- Ulloa L, Batliwalla FM, Andersson U, Gregersen PK, Tracey KJ. High mobility group box chromosomal protein 1 as a nuclear protein, cytokine, and potential therapeutic target in arthritis. Arthritis Rheum. 2003;48:876–81. - PubMed
-
- Czura CJ, Yang H, Amella CA, Tracey KJ. HMGB1 in the immunology of sepsis (not septic shock) and arthritis. Adv Immunol. 2004;84:181–200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

