The promoter of filamentation (POF1) protein from Saccharomyces cerevisiae is an ATPase involved in the protein quality control process
- PMID: 22204397
- PMCID: PMC3282682
- DOI: 10.1186/1471-2180-11-268
The promoter of filamentation (POF1) protein from Saccharomyces cerevisiae is an ATPase involved in the protein quality control process
Abstract
Background: The gene YCL047C, which has been renamed promoter of filamentation gene (POF1), has recently been described as a cell component involved in yeast filamentous growth. The objective of this work is to understand the molecular and biological function of this gene.
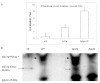
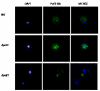
Results: Here, we report that the protein encoded by the POF1 gene, Pof1p, is an ATPase that may be part of the Saccharomyces cerevisiae protein quality control pathway. According to the results, Δpof1 cells showed increased sensitivity to hydrogen peroxide, tert-butyl hydroperoxide, heat shock and protein unfolding agents, such as dithiothreitol and tunicamycin. Besides, the overexpression of POF1 suppressed the sensitivity of Δpct1, a strain that lacks a gene that encodes a phosphocholine cytidylyltransferase, to heat shock. In vitro analysis showed, however, that the purified Pof1p enzyme had no cytidylyltransferase activity but does have ATPase activity, with catalytic efficiency comparable to other ATPases involved in endoplasmic reticulum-associated degradation of proteins (ERAD). Supporting these findings, co-immunoprecipitation experiments showed a physical interaction between Pof1p and Ubc7p (an ubiquitin conjugating enzyme) in vivo.
Conclusions: Taken together, the results strongly suggest that the biological function of Pof1p is related to the regulation of protein degradation.
Figures




References
-
- Leidhold C, Voos W. Chaperones and proteases--guardians of protein integrity in eukaryotic organelles. Ann N Y Acad Sci. 2007. pp. 72–86. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

